Selective selC-independent selenocysteine incorporation into formate dehydrogenases
- PMID: 23634217
- PMCID: PMC3636253
- DOI: 10.1371/journal.pone.0061913
Selective selC-independent selenocysteine incorporation into formate dehydrogenases
Abstract
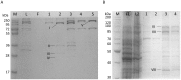
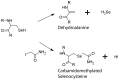
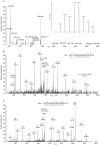
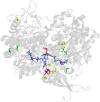
The formate dehydrogenases (Fdh) Fdh-O, Fdh-N, and Fdh-H, are the only proteins in Escherichia coli that incorporate selenocysteine at a specific position by decoding a UGA codon. However, an excess of selenium can lead to toxicity through misincorporation of selenocysteine into proteins. To determine whether selenocysteine substitutes for cysteine, we grew Escherichia coli in the presence of excess sodium selenite. The respiratory Fdh-N and Fdh-O enzymes, along with nitrate reductase (Nar) were co-purified from wild type strain MC4100 after anaerobic growth with nitrate and either 2 µM or 100 µM selenite. Mass spectrometric analysis of the catalytic subunits of both Fdhs identified the UGA-specified selenocysteine residue and revealed incorporation of additional, 'non-specific' selenocysteinyl residues, which always replaced particular cysteinyl residues. Although variable, their incorporation was not random and was independent of the selenite concentration used. Notably, these cysteines are likely to be non-essential for catalysis and they do not coordinate the iron-sulfur cluster. The remaining cysteinyl residues that could be identified were never substituted by selenocysteine. Selenomethionine was never observed in our analyses. Non-random substitution of particular cysteinyl residues was also noted in the electron-transferring subunit of both Fdhs as well as in the subunits of the Nar enzyme. Nar isolated from an E. coli selC mutant also showed a similar selenocysteine incorporation pattern to the wild-type indicating that non-specific selenocysteine incorporation was independent of the specific selenocysteine pathway. Thus, selenide replaces sulfide in the biosynthesis of cysteine and misacylated selenocysteyl-tRNA(Cys) decodes either UGU or UGC codons, which usually specify cysteine. Nevertheless, not every UGU or UGC codon was decoded as selenocysteine. Together, our results suggest that a degree of misincorporation of selenocysteine into enzymes through replacement of particular, non-essential cysteines, is tolerated and this might act as a buffering system to cope with excessive intracellular selenium.
Conflict of interest statement
Figures

Similar articles
-
Effect of the relative position of the UGA codon to the unique secondary structure in the fdhF mRNA on its decoding by selenocysteinyl tRNA in Escherichia coli.J Biol Chem. 1993 Nov 5;268(31):23128-31. J Biol Chem. 1993. PMID: 8226830
-
Molecular and biochemical characterization of two tungsten- and selenium-containing formate dehydrogenases from Eubacterium acidaminophilum that are associated with components of an iron-only hydrogenase.Arch Microbiol. 2003 Jan-Feb;179(2):116-30. doi: 10.1007/s00203-002-0508-1. Epub 2003 Jan 17. Arch Microbiol. 2003. PMID: 12560990
-
Cotranslational insertion of selenocysteine into formate dehydrogenase from Escherichia coli directed by a UGA codon.Proc Natl Acad Sci U S A. 1987 May;84(10):3156-60. doi: 10.1073/pnas.84.10.3156. Proc Natl Acad Sci U S A. 1987. PMID: 3033637 Free PMC article.
-
Selenocysteine.Annu Rev Biochem. 1996;65:83-100. doi: 10.1146/annurev.bi.65.070196.000503. Annu Rev Biochem. 1996. PMID: 8811175 Review.
-
Specific occurrence of selenium in enzymes and amino acid tRNAs.FASEB J. 1987 Nov;1(5):375-9. doi: 10.1096/fasebj.1.5.2445614. FASEB J. 1987. PMID: 2445614 Review.
Cited by
-
Exploring the selenium-over-sulfur substrate specificity and kinetics of a bacterial selenocysteine lyase.Biochimie. 2021 Mar;182:166-176. doi: 10.1016/j.biochi.2021.01.002. Epub 2021 Jan 11. Biochimie. 2021. PMID: 33444662 Free PMC article.
-
Stable and Potent Selenomab-Drug Conjugates.Cell Chem Biol. 2017 Apr 20;24(4):433-442.e6. doi: 10.1016/j.chembiol.2017.02.012. Epub 2017 Mar 16. Cell Chem Biol. 2017. PMID: 28330604 Free PMC article.
-
Phylogeny, distribution and potential metabolism of candidate bacterial phylum KSB1.PeerJ. 2022 Apr 12;10:e13241. doi: 10.7717/peerj.13241. eCollection 2022. PeerJ. 2022. PMID: 35433121 Free PMC article.
-
Metabolic response of Pseudomonas putida to increased NADH regeneration rates.Eng Life Sci. 2016 Oct 4;17(1):47-57. doi: 10.1002/elsc.201600072. eCollection 2017 Jan. Eng Life Sci. 2016. PMID: 32624728 Free PMC article.
-
Exposure to selenomethionine causes selenocysteine misincorporation and protein aggregation in Saccharomyces cerevisiae.Sci Rep. 2017 Mar 17;7:44761. doi: 10.1038/srep44761. Sci Rep. 2017. PMID: 28303947 Free PMC article.
References
-
- Böck A, Forchhammer K, Heider J, Leinfelder W, Sawers G, et al. (1991) Selenocysteine: the 21st amino acid. Mol Microbiol 5: 515–520. - PubMed
-
- Yoshikawa S, Böck A (2009) The many levels of control on bacterial selenoprotein synthesis. Biochim Biophys Acta 1790: 1404–1414. - PubMed
-
- Stadtman TC (1996) Selenocysteine. Ann Rev Biochem 65: 83–100. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

