Maturation of lymph node fibroblastic reticular cells from myofibroblastic precursors is critical for antiviral immunity
- PMID: 23623380
- PMCID: PMC7111182
- DOI: 10.1016/j.immuni.2013.03.012
Maturation of lymph node fibroblastic reticular cells from myofibroblastic precursors is critical for antiviral immunity
Abstract
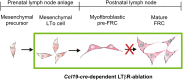
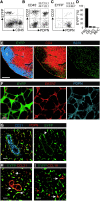
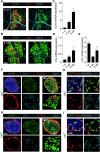
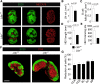
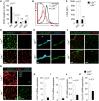
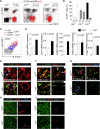
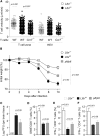
The stromal scaffold of the lymph node (LN) paracortex is built by fibroblastic reticular cells (FRCs). Conditional ablation of lymphotoxin-β receptor (LTβR) expression in LN FRCs and their mesenchymal progenitors in developing LNs revealed that LTβR-signaling in these cells was not essential for the formation of LNs. Although T cell zone reticular cells had lost podoplanin expression, they still formed a functional conduit system and showed enhanced expression of myofibroblastic markers. However, essential immune functions of FRCs, including homeostatic chemokine and interleukin-7 expression, were impaired. These changes in T cell zone reticular cell function were associated with increased susceptibility to viral infection. Thus, myofibroblasic FRC precursors are able to generate the basic T cell zone infrastructure, whereas LTβR-dependent maturation of FRCs guarantees full immunocompetence and hence optimal LN function during infection.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures
Comment in
-
Orchestrating the organizers: lymphotoxin-β receptor conducts fibroblastic reticular cell maturation.Immunity. 2013 May 23;38(5):851-3. doi: 10.1016/j.immuni.2013.05.003. Immunity. 2013. PMID: 23706665 No abstract available.
Similar articles
-
YAP/TAZ direct commitment and maturation of lymph node fibroblastic reticular cells.Nat Commun. 2020 Jan 24;11(1):519. doi: 10.1038/s41467-020-14293-1. Nat Commun. 2020. PMID: 31980640 Free PMC article.
-
SIRPα+ dendritic cells promote the development of fibroblastic reticular cells in murine peripheral lymph nodes.Eur J Immunol. 2019 Sep;49(9):1364-1371. doi: 10.1002/eji.201948103. Epub 2019 Jun 3. Eur J Immunol. 2019. PMID: 31099900
-
Orchestrating the organizers: lymphotoxin-β receptor conducts fibroblastic reticular cell maturation.Immunity. 2013 May 23;38(5):851-3. doi: 10.1016/j.immuni.2013.05.003. Immunity. 2013. PMID: 23706665 No abstract available.
-
Lymph node fibroblastic reticular cells in health and disease.Nat Rev Immunol. 2015 Jun;15(6):350-61. doi: 10.1038/nri3846. Nat Rev Immunol. 2015. PMID: 25998961 Free PMC article. Review.
-
Lymph node fibroblastic reticular cells steer immune responses.Trends Immunol. 2021 Aug;42(8):723-734. doi: 10.1016/j.it.2021.06.006. Epub 2021 Jul 10. Trends Immunol. 2021. PMID: 34256989 Free PMC article. Review.
Cited by
-
How Follicular Dendritic Cells Shape the B-Cell Antigenome.Front Immunol. 2016 Jun 21;7:225. doi: 10.3389/fimmu.2016.00225. eCollection 2016. Front Immunol. 2016. PMID: 27446069 Free PMC article. Review.
-
KDM6B drives epigenetic reprogramming associated with lymphoid stromal cell early commitment and immune properties.Sci Adv. 2023 Dec;9(48):eadh2708. doi: 10.1126/sciadv.adh2708. Epub 2023 Nov 29. Sci Adv. 2023. PMID: 38019914 Free PMC article.
-
The HIV-1 envelope protein gp120 is captured and displayed for B cell recognition by SIGN-R1(+) lymph node macrophages.Elife. 2015 Aug 10;4:e06467. doi: 10.7554/eLife.06467. Elife. 2015. PMID: 26258881 Free PMC article.
-
An essential role for RGS protein/Gαi2 interactions in B lymphocyte-directed cell migration and trafficking.J Immunol. 2015 Mar 1;194(5):2128-39. doi: 10.4049/jimmunol.1401952. Epub 2015 Jan 23. J Immunol. 2015. PMID: 25617475 Free PMC article.
-
Adventitial Stromal Cells Define Group 2 Innate Lymphoid Cell Tissue Niches.Immunity. 2019 Mar 19;50(3):707-722.e6. doi: 10.1016/j.immuni.2019.02.002. Epub 2019 Feb 26. Immunity. 2019. PMID: 30824323 Free PMC article.
References
-
- Adachi S., Yoshida H., Honda K., Maki K., Saijo K., Ikuta K., Saito T., Nishikawa S.I. Essential role of IL-7 receptor alpha in the formation of Peyer’s patch anlage. Int. Immunol. 1998;10:1–6. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

