Reconstitution of hemisomes on budding yeast centromeric DNA
- PMID: 23620291
- PMCID: PMC3675498
- DOI: 10.1093/nar/gkt314
Reconstitution of hemisomes on budding yeast centromeric DNA
Abstract
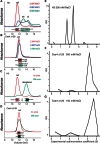
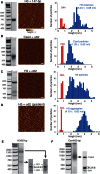
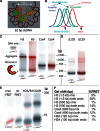
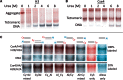
The structure of nucleosomes that contain the cenH3 histone variant has been controversial. In budding yeast, a single right-handed cenH3/H4/H2A/H2B tetramer wraps the ∼80-bp Centromere DNA Element II (CDE II) sequence of each centromere into a 'hemisome'. However, attempts to reconstitute cenH3 particles in vitro have yielded exclusively 'octasomes', which are observed in vivo on chromosome arms only when Cse4 (yeast cenH3) is overproduced. Here, we show that Cse4 octamers remain intact under conditions of low salt and urea that dissociate H3 octamers. However, particles consisting of two DNA duplexes wrapped around a Cse4 octamer and separated by a gap efficiently split into hemisomes. Hemisome dimensions were confirmed using a calibrated gel-shift assay and atomic force microscopy, and their identity as tightly wrapped particles was demonstrated by gelFRET. Surprisingly, Cse4 hemisomes were stable in 4 M urea. Stable Cse4 hemisomes could be reconstituted using either full-length or tailless histones and with a 78-bp CDEII segment, which is predicted to be exceptionally stiff. We propose that CDEII DNA stiffness evolved to favor Cse4 hemisome over octasome formation. The precise correspondence between Cse4 hemisomes resident on CDEII in vivo and reconstituted on CDEII in vitro without any other factors implies that CDEII is sufficient for hemisome assembly.
Figures

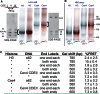
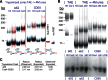
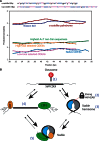
Similar articles
-
The budding yeast Centromere DNA Element II wraps a stable Cse4 hemisome in either orientation in vivo.Elife. 2014 Apr 15;3:e01861. doi: 10.7554/eLife.01861. Elife. 2014. PMID: 24737863 Free PMC article.
-
The unconventional structure of centromeric nucleosomes.Chromosoma. 2012 Aug;121(4):341-52. doi: 10.1007/s00412-012-0372-y. Epub 2012 May 3. Chromosoma. 2012. PMID: 22552438 Free PMC article. Review.
-
Nonhistone Scm3 binds to AT-rich DNA to organize atypical centromeric nucleosome of budding yeast.Mol Cell. 2011 Aug 5;43(3):369-80. doi: 10.1016/j.molcel.2011.07.009. Mol Cell. 2011. PMID: 21816344 Free PMC article.
-
Structural basis for recognition of centromere histone variant CenH3 by the chaperone Scm3.Nature. 2011 Apr 14;472(7342):234-7. doi: 10.1038/nature09854. Epub 2011 Mar 16. Nature. 2011. PMID: 21412236 Free PMC article.
-
Structure, dynamics, and evolution of centromeric nucleosomes.Proc Natl Acad Sci U S A. 2007 Oct 9;104(41):15974-81. doi: 10.1073/pnas.0707648104. Epub 2007 Sep 24. Proc Natl Acad Sci U S A. 2007. PMID: 17893333 Free PMC article. Review.
Cited by
-
Structure of the human inner kinetochore CCAN complex and its significance for human centromere organization.Mol Cell. 2022 Jun 2;82(11):2113-2131.e8. doi: 10.1016/j.molcel.2022.04.027. Epub 2022 May 6. Mol Cell. 2022. PMID: 35525244 Free PMC article.
-
Reply to "CENP-A octamers do not confer a reduction in nucleosome height by AFM".Nat Struct Mol Biol. 2014 Jan;21(1):5-8. doi: 10.1038/nsmb.2744. Nat Struct Mol Biol. 2014. PMID: 24389543 Free PMC article. No abstract available.
-
Imaging the fate of histone Cse4 reveals de novo replacement in S phase and subsequent stable residence at centromeres.Elife. 2014 May 20;3:e02203. doi: 10.7554/eLife.02203. Elife. 2014. PMID: 24844245 Free PMC article.
-
Shearing of the CENP-A dimerization interface mediates plasticity in the octameric centromeric nucleosome.Sci Rep. 2015 Nov 25;5:17038. doi: 10.1038/srep17038. Sci Rep. 2015. PMID: 26602160 Free PMC article.
-
Novel genetic tools for probing individual H3 molecules in each nucleosome.Curr Genet. 2019 Apr;65(2):371-377. doi: 10.1007/s00294-018-0910-0. Epub 2018 Nov 26. Curr Genet. 2019. PMID: 30478690 Free PMC article. Review.
References
-
- Malik HS, Henikoff S. Major evolutionary transitions in centromere complexity. Cell. 2009;138:1067–1082. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

