Lumenal peroxisomal protein aggregates are removed by concerted fission and autophagy events
- PMID: 23614977
- PMCID: PMC3722314
- DOI: 10.4161/auto.24543
Lumenal peroxisomal protein aggregates are removed by concerted fission and autophagy events
Abstract
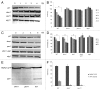
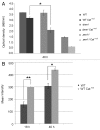
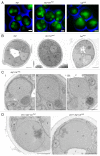
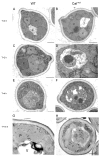
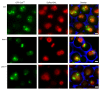
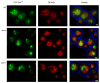
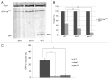
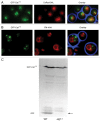
We demonstrated that in the yeast Hansenula polymorpha peroxisome fission and degradation are coupled processes that are important to remove intra-organellar protein aggregates. Protein aggregates were formed in peroxisomes upon synthesis of a mutant catalase variant. We showed that the introduction of these aggregates in the peroxisomal lumen had physiological disadvantages as it affected growth and caused enhanced levels of reactive oxygen species. Formation of the protein aggregates was followed by asymmetric peroxisome fission to separate the aggregate from the mother organelle. Subsequently, these small, protein aggregate-containing organelles were degraded by autophagy. In line with this observation we showed that the degradation of the protein aggregates was strongly reduced in dnm1 and pex11 cells in which peroxisome fission is reduced. Moreover, this process was dependent on Atg1 and Atg11.
Keywords: autophagy; fission; peroxisome; protein aggregate; yeast.
Figures
Similar articles
-
A peroxisomal lon protease and peroxisome degradation by autophagy play key roles in vitality of Hansenula polymorpha cells.Autophagy. 2007 Mar-Apr;3(2):96-105. doi: 10.4161/auto.3534. Epub 2007 Mar 23. Autophagy. 2007. PMID: 17172804
-
Peroxisome fission in Hansenula polymorpha requires Mdv1 and Fis1, two proteins also involved in mitochondrial fission.Traffic. 2008 Sep;9(9):1471-84. doi: 10.1111/j.1600-0854.2008.00772.x. Epub 2008 May 30. Traffic. 2008. PMID: 18513378
-
Damaged peroxisomes are subject to rapid autophagic degradation in the yeast Hansenula polymorpha.Autophagy. 2011 Aug;7(8):863-72. doi: 10.4161/auto.7.8.15697. Epub 2011 Aug 1. Autophagy. 2011. PMID: 21490428
-
Peroxisome homeostasis in Hansenula polymorpha.FEMS Yeast Res. 2003 Nov;4(2):131-9. doi: 10.1016/S1567-1356(03)00070-9. FEMS Yeast Res. 2003. PMID: 14613877 Review.
-
Peroxisomal matrix protein import. Suppression of protein import defects in Hansenula polymorpha pex mutants by overproduction of the PTS1 receptor Pex5p.Cell Biochem Biophys. 2000;32 Spring:9-19. doi: 10.1385/cbb:32:1-3:09. Cell Biochem Biophys. 2000. PMID: 11330074 Review.
Cited by
-
Mechanisms and Functions of Pexophagy in Mammalian Cells.Cells. 2021 May 3;10(5):1094. doi: 10.3390/cells10051094. Cells. 2021. PMID: 34063724 Free PMC article. Review.
-
The different facets of organelle interplay-an overview of organelle interactions.Front Cell Dev Biol. 2015 Sep 25;3:56. doi: 10.3389/fcell.2015.00056. eCollection 2015. Front Cell Dev Biol. 2015. PMID: 26442263 Free PMC article. Review.
-
Hansenula polymorpha Pex37 is a peroxisomal membrane protein required for organelle fission and segregation.FEBS J. 2020 May;287(9):1742-1757. doi: 10.1111/febs.15123. Epub 2019 Nov 26. FEBS J. 2020. PMID: 31692262 Free PMC article.
-
Distinct Contributions of the Peroxisome-Mitochondria Fission Machinery During Sexual Development of the Fungus Podospora anserina.Front Microbiol. 2020 Apr 15;11:640. doi: 10.3389/fmicb.2020.00640. eCollection 2020. Front Microbiol. 2020. PMID: 32351478 Free PMC article.
-
Organellophagy: eliminating cellular building blocks via selective autophagy.J Cell Biol. 2014 May 26;205(4):435-45. doi: 10.1083/jcb.201402054. J Cell Biol. 2014. PMID: 24862571 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
