In vivo analysis of Aicda gene regulation: a critical balance between upstream enhancers and intronic silencers governs appropriate expression
- PMID: 23613851
- PMCID: PMC3628980
- DOI: 10.1371/journal.pone.0061433
In vivo analysis of Aicda gene regulation: a critical balance between upstream enhancers and intronic silencers governs appropriate expression
Abstract
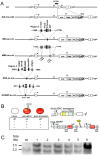
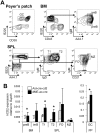
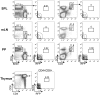
The Aicda gene encodes activation-induced cytidine deaminase (AID). Aicda is strongly transcribed in activated B cells to diversify immunoglobulin genes, but expressed at low levels in various other cells in response to physiological or pathological stimuli. AID's mutagenic nature has been shown to be involved in tumor development. Here, we used a transgenic strategy with bacterial artificial chromosomes (BACs) to examine the in vivo functions of Aicda regulatory elements, which cluster in two regions: in the first intron (region 2), and approximately 8-kb upstream of the transcription start site (region 4). Deleting either of these regions completely abolished the expression of Aicda-BAC reporters, demonstrating these elements' critical roles. Furthermore, we found that selectively deleting two C/EBP-binding sites in region 4 inactivated the enhancer activity of the region despite the presence of intact NF-κB-, STAT6- and Smad-binding sites. On the other hand, selectively deleting E2F- and c-Myb-binding sites in region 2 increased the frequency of germinal-center B cells in which the Aicda promoter was active, indicating that E2F and c-Myb act as silencers in vivo. Interestingly, the silencer deletion did not cause ectopic activation of the Aicda promoter, indicating that Aicda activation requires enhancer-specific stimulation. In summary, precise regulation of the Aicda promoter appears to depend on a coordinated balance of activities between enhancer and silencer elements.
Conflict of interest statement
Figures



Similar articles
-
B cell-specific and stimulation-responsive enhancers derepress Aicda by overcoming the effects of silencers.Nat Immunol. 2010 Feb;11(2):148-54. doi: 10.1038/ni.1829. Epub 2009 Dec 6. Nat Immunol. 2010. PMID: 19966806
-
Identification of a ubiquitously active promoter of the murine activation-induced cytidine deaminase (AICDA) gene.Mol Immunol. 2006 Feb;43(6):529-41. doi: 10.1016/j.molimm.2005.05.007. Epub 2005 Jul 6. Mol Immunol. 2006. PMID: 16005067
-
Transcriptional regulation of teleost Aicda genes. Part 1 - suppressors of promiscuous promoters.Fish Shellfish Immunol. 2013 Dec;35(6):1981-7. doi: 10.1016/j.fsi.2013.09.035. Epub 2013 Oct 24. Fish Shellfish Immunol. 2013. PMID: 24161771
-
Regulatory regions in DNA: promoters, enhancers, silencers, and insulators.Methods Mol Biol. 2010;674:33-42. doi: 10.1007/978-1-60761-854-6_3. Methods Mol Biol. 2010. PMID: 20827584 Review.
-
The current research status of enhancer RNAs.Yi Chuan. 2017 Sep 20;39(9):784-797. doi: 10.16288/j.yczz.17-010. Yi Chuan. 2017. PMID: 28936977 Review.
Cited by
-
MicroRNA binding site polymorphism in inflammatory genes associated with colorectal cancer: literature review and bioinformatics analysis.Cancer Gene Ther. 2020 Nov;27(10-11):739-753. doi: 10.1038/s41417-020-0172-0. Epub 2020 Mar 23. Cancer Gene Ther. 2020. PMID: 32203060 Review.
-
Regulation of immunoglobulin class-switch recombination: choreography of noncoding transcription, targeted DNA deamination, and long-range DNA repair.Adv Immunol. 2014;122:1-57. doi: 10.1016/B978-0-12-800267-4.00001-8. Adv Immunol. 2014. PMID: 24507154 Free PMC article. Review.
-
Mutations, kataegis and translocations in B cells: understanding AID promiscuous activity.Nat Rev Immunol. 2016 Mar;16(3):164-76. doi: 10.1038/nri.2016.2. Epub 2016 Feb 22. Nat Rev Immunol. 2016. PMID: 26898111 Free PMC article. Review.
-
Suppression by TFR cells leads to durable and selective inhibition of B cell effector function.Nat Immunol. 2016 Dec;17(12):1436-1446. doi: 10.1038/ni.3578. Epub 2016 Oct 3. Nat Immunol. 2016. PMID: 27695002 Free PMC article.
-
Molecular functions of the transcription factors E2A and E2-2 in controlling germinal center B cell and plasma cell development.J Exp Med. 2016 Jun 27;213(7):1201-21. doi: 10.1084/jem.20152002. Epub 2016 Jun 3. J Exp Med. 2016. PMID: 27261530 Free PMC article.
References
-
- Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, et al. (2000) Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell 102: 553–563. - PubMed
-
- Revy P, Muto T, Levy Y, Geissmann F, Plebani A, et al. (2000) Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2). Cell 102: 565–575. - PubMed
-
- Liu M, Duke JL, Richter DJ, Vinuesa CG, Goodnow CC, et al. (2008) Two levels of protection for the B cell genome during somatic hypermutation. Nature 451: 841–845. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

