New synthetic substrates of mammalian nucleotide excision repair system
- PMID: 23609543
- PMCID: PMC3695498
- DOI: 10.1093/nar/gkt301
New synthetic substrates of mammalian nucleotide excision repair system
Abstract
DNA probes for the studies of damaged strand excision during the nucleotide excision repair (NER) have been designed using the novel non-nucleosidic phosphoramidite reagents that contain N-[6-(9-antracenylcarbamoyl)hexanoyl]-3-amino-1,2-propandiol (nAnt) and N-[6-(5(6)-fluoresceinylcarbamoyl)hexanoyl]-3-amino-1,2-propandiol (nFlu) moieties. New lesion-imitating adducts being inserted into DNA show good substrate properties in NER process. Modified extended linear nFlu- and nAntr-DNA are suitable for estimation of specific excision activity catalysed with mammalian whole-cell extracts. The following substrate activity range was revealed for the model 137-bp linear double-stranded DNA: nAnt-DNA ≈ nFlu-DNA > Chol-DNA (Chol-DNA--legitimate NER substrate that contains non-nucleoside fragment bearing cholesterol residue). In vitro assay shows that modified DNA can be a useful tool to study NER activity in whole-cell extracts. The developed approach should be of general use for the incorporation of NER-sensitive distortions into model DNAs. The new synthetic extended linear DNA containing bulky non-nucleoside modifications will be useful for NER mechanism study and for applications.
Figures
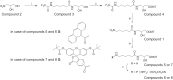
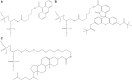
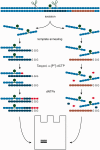


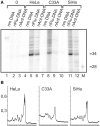
Similar articles
-
[DNA Bearing Bulky Fluorescent and Photoreactive Damage in Both Strands as Substrates of the Nucleotide Excision Repair System].Mol Biol (Mosk). 2018 Mar-Apr;52(2):277-288. doi: 10.7868/S0026898418020118. Mol Biol (Mosk). 2018. PMID: 29695696 Russian.
-
DNA with Damage in Both Strands as Affinity Probes and Nucleotide Excision Repair Substrates.Biochemistry (Mosc). 2016 Mar;81(3):263-74. doi: 10.1134/S0006297916030093. Biochemistry (Mosc). 2016. PMID: 27262196
-
End modification of a linear DNA duplex enhances NER-mediated excision of an internal Pt(II)-lesion.Bioconjug Chem. 2008 May;19(5):1064-70. doi: 10.1021/bc7004363. Epub 2008 May 1. Bioconjug Chem. 2008. PMID: 18447369
-
Eukaryotic nucleotide excision repair: from understanding mechanisms to influencing biology.Cell Res. 2008 Jan;18(1):64-72. doi: 10.1038/cr.2008.2. Cell Res. 2008. PMID: 18166981 Free PMC article. Review.
-
Emerging roles for histone modifications in DNA excision repair.FEMS Yeast Res. 2016 Nov;16(7):fow090. doi: 10.1093/femsyr/fow090. Epub 2016 Oct 12. FEMS Yeast Res. 2016. PMID: 27737893 Free PMC article. Review.
Cited by
-
Naked mole rat cells display more efficient excision repair than mouse cells.Aging (Albany NY). 2018 Jun 20;10(6):1454-1473. doi: 10.18632/aging.101482. Aging (Albany NY). 2018. PMID: 29930219 Free PMC article.
-
The Interaction Efficiency of XPD-p44 With Bulky DNA Damages Depends on the Structure of the Damage.Front Cell Dev Biol. 2021 Mar 11;9:617160. doi: 10.3389/fcell.2021.617160. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 33777931 Free PMC article.
-
Fluorescence detection of cellular nucleotide excision repair of damaged DNA.Sci Rep. 2014 Jul 4;4:5578. doi: 10.1038/srep05578. Sci Rep. 2014. PMID: 24993089 Free PMC article.
-
Cholesterol Conjugates of Small Interfering RNA: Linkers and Patterns of Modification.Molecules. 2024 Feb 8;29(4):786. doi: 10.3390/molecules29040786. Molecules. 2024. PMID: 38398538 Free PMC article.
-
A Method for Assessing the Efficiency of the Nucleotide Excision Repair System Ex Vivo.Acta Naturae. 2021 Jul-Sep;13(3):122-125. doi: 10.32607/actanaturae.11430. Acta Naturae. 2021. PMID: 34707905 Free PMC article.
References
-
- Gillet LC, Scharer OD. Molecular mechanisms of mammalian global genome nucleotide excision repair. Chem. Rev. 2006;106:253–276. - PubMed
-
- Huang JC, Sancar A. Determination of minimum substrate size for human excinuclease. J. Biol. Chem. 1994;269:19034–19040. - PubMed
-
- Geacintov NE, Broyde S, Buterin T, Naegeli H, Wu M, Yan S, Patel DJ. Thermodynamic and structural factors in the removal of bulky DNA adducts by the nucleotide excision repair machinery. Biopolymers. 2002;65:202–210. - PubMed
-
- DellaVecchia MJ, Croteau DL, Skorvaga M, Dezhurov SV, Lavrik OI, Van Houten B. Analyzing the handoff of DNA from UvrA to UvrB utilizing DNA-protein photoaffinity labeling. J. Biol. Chem. 2004;279:45245–45256. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

