Distinct binding properties of TIAR RRMs and linker region
- PMID: 23603827
- PMCID: PMC3710364
- DOI: 10.4161/rna.24341
Distinct binding properties of TIAR RRMs and linker region
Abstract
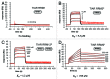
The RNA-binding protein TIAR is an mRNA-binding protein that acts as a translational repressor, particularly important under conditions of cellular stress. It binds to target mRNA and DNA via its RNA recognition motif (RRM) domains and is involved in both splicing regulation and translational repression via the formation of "stress granules." TIAR has also been shown to bind ssDNA and play a role in the regulation of transcription. Here we show, using surface plasmon resonance and nuclear magnetic resonance spectroscopy, specific roles of individual TIAR domains for high-affinity binding to RNA and DNA targets. We confirm that RRM2 of TIAR is the major RNA- and DNA-binding domain. However, the strong nanomolar affinity binding to U-rich RNA and T-rich DNA depends on the presence of the six amino acid residues found in the linker region C-terminal to RRM2. On its own, RRM1 shows preferred binding to DNA over RNA. We further characterize the interaction between RRM2 with the C-terminal extension and an AU-rich target RNA sequence using NMR spectroscopy to identify the amino acid residues involved in binding. We demonstrate that TIAR RRM2, together with its C-terminal extension, is the major contributor for the high-affinity (nM) interactions of TIAR with target RNA sequences.
Keywords: C-terminal extension; NMR; RNA-binding protein; RRM; TIA-1; TIAR; surface plasmon resonance (SPR); translational regulation.
Figures


Similar articles
-
The C-terminal RNA binding motif of HuR is a multi-functional domain leading to HuR oligomerization and binding to U-rich RNA targets.RNA Biol. 2014;11(10):1250-61. doi: 10.1080/15476286.2014.996069. RNA Biol. 2014. PMID: 25584704 Free PMC article.
-
Different modes of interaction by TIAR and HuR with target RNA and DNA.Nucleic Acids Res. 2011 Feb;39(3):1117-30. doi: 10.1093/nar/gkq837. Epub 2011 Jan 13. Nucleic Acids Res. 2011. PMID: 21233170 Free PMC article.
-
Elucidation of a C-rich signature motif in target mRNAs of RNA-binding protein TIAR.Mol Cell Biol. 2007 Oct;27(19):6806-17. doi: 10.1128/MCB.01036-07. Epub 2007 Aug 6. Mol Cell Biol. 2007. PMID: 17682065 Free PMC article.
-
AU-rich element-mediated translational control: complexity and multiple activities of trans-activating factors.Biochem Soc Trans. 2002 Nov;30(Pt 6):952-8. doi: 10.1042/bst0300952. Biochem Soc Trans. 2002. PMID: 12440953 Review.
-
RNA recognition and stress granule formation by TIA proteins.Int J Mol Sci. 2014 Dec 16;15(12):23377-88. doi: 10.3390/ijms151223377. Int J Mol Sci. 2014. PMID: 25522169 Free PMC article. Review.
Cited by
-
Balanced splicing at the Tat-specific HIV-1 3'ss A3 is critical for HIV-1 replication.Retrovirology. 2015 Mar 28;12:29. doi: 10.1186/s12977-015-0154-8. Retrovirology. 2015. PMID: 25889056 Free PMC article.
-
T-Cell Intracellular Antigen 1-Like Protein in Physiology and Pathology.Int J Mol Sci. 2022 Jul 16;23(14):7836. doi: 10.3390/ijms23147836. Int J Mol Sci. 2022. PMID: 35887183 Free PMC article. Review.
-
Tandem RNA binding sites induce self-association of the stress granule marker protein TIA-1.Nucleic Acids Res. 2021 Mar 18;49(5):2403-2417. doi: 10.1093/nar/gkab080. Nucleic Acids Res. 2021. PMID: 33621982 Free PMC article.
-
Role of 3'-untranslated region translational control in cancer development, diagnostics and treatment.World J Biol Chem. 2014 Feb 26;5(1):40-57. doi: 10.4331/wjbc.v5.i1.40. World J Biol Chem. 2014. PMID: 24600513 Free PMC article. Review.
-
Posttranscriptional Regulation of Intestinal Epithelial Tight Junction Barrier by RNA-binding Proteins and microRNAs.Tissue Barriers. 2014 Jan 1;2(1):e28320. doi: 10.4161/tisb.28320. Epub 2014 Mar 19. Tissue Barriers. 2014. PMID: 24843843 Free PMC article. Review.
References
-
- Aznarez I, Barash Y, Shai O, He D, Zielenski J, Tsui LC, et al. A systematic analysis of intronic sequences downstream of 5′ splice sites reveals a widespread role for U-rich motifs and TIA1/TIAL1 proteins in alternative splicing regulation. Genome Res. 2008;18:1247–58. doi: 10.1101/gr.073155.107. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
