Replication stress induces specific enrichment of RECQ1 at common fragile sites FRA3B and FRA16D
- PMID: 23601052
- PMCID: PMC3663727
- DOI: 10.1186/1476-4598-12-29
Replication stress induces specific enrichment of RECQ1 at common fragile sites FRA3B and FRA16D
Abstract
Background: Stalled replication forks at common fragile sites are a major cause of genomic instability. RecQ helicases, a highly conserved family of DNA-unwinding enzymes, are believed to ease 'roadblocks' that pose challenge to replication fork progression. Among the five known RecQ homologs in humans, functions of RECQ1, the most abundant of all, are poorly understood. We previously determined that RECQ1 helicase preferentially binds and unwinds substrates that mimic DNA replication/repair intermediates, and interacts with proteins involved in DNA replication restart mechanisms.
Method: We have utilized chromatin immunoprecipitation followed by quantitative real-time PCR to investigate chromatin interactions of RECQ1 at defined genetic loci in the presence or absence of replication stress. We have also tested the sensitivity of RECQ1-depleted cells to aphidicolin induced replication stress.
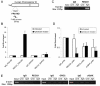
Results: RECQ1 binds to the origins of replication in unperturbed cells. We now show that conditions of replication stress induce increased accumulation of RECQ1 at the lamin B2 origin in HeLa cells. Consistent with a role in promoting fork recovery or repair, RECQ1 is specifically enriched at two major fragile sites FRA3B and FRA16D where replication forks have stalled following aphidicolin treatment. RECQ1-depletion results in attenuated checkpoint activation in response to replication stress, increased sensitivity to aphidicolin and chromosomal instability.
Conclusions: Given a recent biochemical observation that RECQ1 catalyzes strand exchange on stalled replication fork structures in vitro, our results indicate that RECQ1 facilitates repair of stalled or collapsed replication forks and preserves genome integrity. Our findings provide the first evidence of a crucial role for RECQ1 at naturally occurring fork stalling sites and implicate RECQ1 in mechanisms underlying common fragile site instability in cancer.
Figures



Similar articles
-
RECQ1 is required for cellular resistance to replication stress and catalyzes strand exchange on stalled replication fork structures.Cell Cycle. 2012 Nov 15;11(22):4252-65. doi: 10.4161/cc.22581. Epub 2012 Oct 24. Cell Cycle. 2012. PMID: 23095637 Free PMC article.
-
RECQ1 interacts with FEN-1 and promotes binding of FEN-1 to telomeric chromatin.Biochem J. 2015 Jun 1;468(2):227-44. doi: 10.1042/BJ20141021. Biochem J. 2015. PMID: 25774876 Free PMC article.
-
Catalytic strand separation by RECQ1 is required for RPA-mediated response to replication stress.Curr Biol. 2015 Nov 2;25(21):2830-2838. doi: 10.1016/j.cub.2015.09.026. Epub 2015 Oct 8. Curr Biol. 2015. PMID: 26455304 Free PMC article.
-
Distinct roles of RECQ1 in the maintenance of genomic stability.DNA Repair (Amst). 2010 Mar 2;9(3):315-24. doi: 10.1016/j.dnarep.2009.12.010. Epub 2010 Jan 12. DNA Repair (Amst). 2010. PMID: 20061189 Free PMC article. Review.
-
Distinct functions of human RecQ helicases during DNA replication.Biophys Chem. 2017 Jun;225:20-26. doi: 10.1016/j.bpc.2016.11.005. Epub 2016 Nov 15. Biophys Chem. 2017. PMID: 27876204 Review.
Cited by
-
An appraisal of RECQ1 expression in cancer progression.Front Genet. 2014 Dec 5;5:426. doi: 10.3389/fgene.2014.00426. eCollection 2014. Front Genet. 2014. PMID: 25538733 Free PMC article.
-
RECQ1 expression is upregulated in response to DNA damage and in a p53-dependent manner.Oncotarget. 2017 May 27;8(44):75924-75942. doi: 10.18632/oncotarget.18237. eCollection 2017 Sep 29. Oncotarget. 2017. PMID: 29100281 Free PMC article.
-
Transcriptome guided identification of novel functions of RECQ1 helicase.Methods. 2016 Oct 1;108:111-7. doi: 10.1016/j.ymeth.2016.04.018. Epub 2016 Apr 18. Methods. 2016. PMID: 27102625 Free PMC article.
-
Probing Genome Maintenance Functions of human RECQ1.Comput Struct Biotechnol J. 2013 Oct 18;6:e201303014. doi: 10.5936/csbj.201303014. eCollection 2013. Comput Struct Biotechnol J. 2013. PMID: 24688722 Free PMC article. Review.
-
Replication fork recovery and regulation of common fragile sites stability.Cell Mol Life Sci. 2014 Dec;71(23):4507-17. doi: 10.1007/s00018-014-1718-9. Epub 2014 Sep 13. Cell Mol Life Sci. 2014. PMID: 25216703 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

