Evolutionary dynamics of retrotransposons assessed by high-throughput sequencing in wild relatives of wheat
- PMID: 23595021
- PMCID: PMC4104650
- DOI: 10.1093/gbe/evt064
Evolutionary dynamics of retrotransposons assessed by high-throughput sequencing in wild relatives of wheat
Abstract
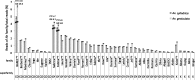
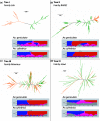
Transposable elements (TEs) represent a major fraction of plant genomes and drive their evolution. An improved understanding of genome evolution requires the dynamics of a large number of TE families to be considered. We put forward an approach bypassing the required step of a complete reference genome to assess the evolutionary trajectories of high copy number TE families from genome snapshot with high-throughput sequencing. Low coverage sequencing of the complex genomes of Aegilops cylindrica and Ae. geniculata using 454 identified more than 70% of the sequences as known TEs, mainly long terminal repeat (LTR) retrotransposons. Comparing the abundance of reads as well as patterns of sequence diversity and divergence within and among genomes assessed the dynamics of 44 major LTR retrotransposon families of the 165 identified. In particular, molecular population genetics on individual TE copies distinguished recently active from quiescent families and highlighted different evolutionary trajectories of retrotransposons among related species. This work presents a suite of tools suitable for current sequencing data, allowing to address the genome-wide evolutionary dynamics of TEs at the family level and advancing our understanding of the evolution of nonmodel genomes.
Keywords: 454 pyrosequencing; Aegilops; molecular population genetics; repetitive fraction composition; transposable elements; whole-genome snapshot.
Figures
Similar articles
-
Falls prevention interventions for community-dwelling older adults: systematic review and meta-analysis of benefits, harms, and patient values and preferences.Syst Rev. 2024 Nov 26;13(1):289. doi: 10.1186/s13643-024-02681-3. Syst Rev. 2024. PMID: 39593159 Free PMC article.
-
Defining the optimum strategy for identifying adults and children with coeliac disease: systematic review and economic modelling.Health Technol Assess. 2022 Oct;26(44):1-310. doi: 10.3310/ZUCE8371. Health Technol Assess. 2022. PMID: 36321689 Free PMC article.
-
Comparison of Two Modern Survival Prediction Tools, SORG-MLA and METSSS, in Patients With Symptomatic Long-bone Metastases Who Underwent Local Treatment With Surgery Followed by Radiotherapy and With Radiotherapy Alone.Clin Orthop Relat Res. 2024 Dec 1;482(12):2193-2208. doi: 10.1097/CORR.0000000000003185. Epub 2024 Jul 23. Clin Orthop Relat Res. 2024. PMID: 39051924
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
-
Impact of residual disease as a prognostic factor for survival in women with advanced epithelial ovarian cancer after primary surgery.Cochrane Database Syst Rev. 2022 Sep 26;9(9):CD015048. doi: 10.1002/14651858.CD015048.pub2. Cochrane Database Syst Rev. 2022. PMID: 36161421 Free PMC article. Review.
Cited by
-
Deep analysis of wild Vitis flower transcriptome reveals unexplored genome regions associated with sex specification.Plant Mol Biol. 2017 Jan;93(1-2):151-170. doi: 10.1007/s11103-016-0553-9. Epub 2016 Oct 24. Plant Mol Biol. 2017. PMID: 27778293
-
Chromosome Specific Substitution Lines of Aegilops geniculata Alter Parameters of Bread Making Quality of Wheat.PLoS One. 2016 Oct 18;11(10):e0162350. doi: 10.1371/journal.pone.0162350. eCollection 2016. PLoS One. 2016. PMID: 27755540 Free PMC article.
-
Sequencing of chloroplast genomes from wheat, barley, rye and their relatives provides a detailed insight into the evolution of the Triticeae tribe.PLoS One. 2014 Mar 10;9(3):e85761. doi: 10.1371/journal.pone.0085761. eCollection 2014. PLoS One. 2014. PMID: 24614886 Free PMC article.
-
Genome reorganization in F1 hybrids uncovers the role of retrotransposons in reproductive isolation.Proc Biol Sci. 2015 Apr 7;282(1804):20142874. doi: 10.1098/rspb.2014.2874. Proc Biol Sci. 2015. PMID: 25716787 Free PMC article.
-
Transposable element dynamics among asymbiotic and ectomycorrhizal Amanita fungi.Genome Biol Evol. 2014 Jun 12;6(7):1564-78. doi: 10.1093/gbe/evu121. Genome Biol Evol. 2014. PMID: 24923322 Free PMC article.
References
-
- Badaeva ED, et al. Genome differentiation in Aegilops. 4. Evolution of the U-genome cluster. Plant Syst Evol. 2004;246:45–76.
-
- Baum BR, et al. Phylogenetic relationships among the polyploid and diploid Aegilops species inferred from the nuclear 5S rDNA sequences (Poaceae: Triticeae) Genome. 2012;55:177–193. - PubMed
-
- Bennetzen JL. Transposable elements, gene creation and genome rearrangement in flowering plants. Curr Opin Genet Dev. 2005;15:621–627. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

