The exonuclease activity of the yeast mitochondrial DNA polymerase γ suppresses mitochondrial DNA deletions between short direct repeats in Saccharomyces cerevisiae
- PMID: 23589460
- PMCID: PMC3664861
- DOI: 10.1534/genetics.113.150920
The exonuclease activity of the yeast mitochondrial DNA polymerase γ suppresses mitochondrial DNA deletions between short direct repeats in Saccharomyces cerevisiae
Abstract
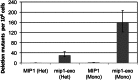
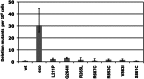
The importance of mitochondrial DNA (mtDNA) deletions in the progeroid phenotype of exonuclease-deficient DNA polymerase γ mice has been intensely debated. We show that disruption of Mip1 exonuclease activity increases mtDNA deletions 160-fold, whereas disease-associated polymerase variants were mostly unaffected, suggesting that exonuclease activity is vital to avoid deletions during mtDNA replication.
Keywords: Mip1; Pol Gamma; direct repeats; exonuclease; mtDNA deletions.
Figures
Similar articles
-
MMS exposure promotes increased MtDNA mutagenesis in the presence of replication-defective disease-associated DNA polymerase γ variants.PLoS Genet. 2014 Oct 23;10(10):e1004748. doi: 10.1371/journal.pgen.1004748. eCollection 2014 Oct. PLoS Genet. 2014. PMID: 25340760 Free PMC article.
-
A non-radioactive DNA synthesis assay demonstrates that elements of the Sigma 1278b Mip1 mitochondrial DNA polymerase domain and C-terminal extension facilitate robust enzyme activity.Yeast. 2021 Apr;38(4):262-275. doi: 10.1002/yea.3541. Epub 2021 Jan 26. Yeast. 2021. PMID: 33270277 Free PMC article.
-
Amino and carboxy-terminal extensions of yeast mitochondrial DNA polymerase assemble both the polymerization and exonuclease active sites.Mitochondrion. 2019 Nov;49:166-177. doi: 10.1016/j.mito.2019.08.005. Epub 2019 Aug 21. Mitochondrion. 2019. PMID: 31445096
-
The Saccharomyces cerevisiae mitochondrial DNA polymerase and its contribution to the knowledge about human POLG-related disorders.IUBMB Life. 2023 Dec;75(12):983-1002. doi: 10.1002/iub.2770. Epub 2023 Jul 20. IUBMB Life. 2023. PMID: 37470284 Review.
-
Mitochondrial DNA mutators.Cell Mol Life Sci. 2004 Nov;61(22):2799-811. doi: 10.1007/s00018-004-4220-y. Cell Mol Life Sci. 2004. PMID: 15558210 Review.
Cited by
-
Saccharomyces cerevisiae as a Tool for Studying Mutations in Nuclear Genes Involved in Diseases Caused by Mitochondrial DNA Instability.Genes (Basel). 2021 Nov 24;12(12):1866. doi: 10.3390/genes12121866. Genes (Basel). 2021. PMID: 34946817 Free PMC article. Review.
-
Yeast model analysis of novel polymerase gamma variants found in patients with autosomal recessive mitochondrial disease.Hum Genet. 2015 Sep;134(9):951-66. doi: 10.1007/s00439-015-1578-x. Epub 2015 Jun 16. Hum Genet. 2015. PMID: 26077851 Free PMC article.
-
DNA polymerase γ and disease: what we have learned from yeast.Front Genet. 2015 Mar 17;6:106. doi: 10.3389/fgene.2015.00106. eCollection 2015. Front Genet. 2015. PMID: 25852747 Free PMC article. Review.
-
Ultrasensitive deletion detection links mitochondrial DNA replication, disease, and aging.Genome Biol. 2020 Sep 17;21(1):248. doi: 10.1186/s13059-020-02138-5. Genome Biol. 2020. PMID: 32943091 Free PMC article.
-
Polymorphisms in DNA polymerase γ affect the mtDNA stability and the NRTI-induced mitochondrial toxicity in Saccharomyces cerevisiae.Mitochondrion. 2015 Jan;20:52-63. doi: 10.1016/j.mito.2014.11.003. Epub 2014 Nov 18. Mitochondrion. 2015. PMID: 25462018 Free PMC article.
References
-
- Baruffini E., Ferrero I., Foury F., 2007. Mitochondrial DNA defects in Saccharomyces cerevisiae caused by functional interactions between DNA polymerase gamma mutations associated with disease in human. Biochim. Biophys. Acta 1772: 1225–1235. - PubMed
-
- Baruffini E., Lodi T., Dallabona C., Puglisi A., Zeviani M., et al. , 2006. Genetic and chemical rescue of the Saccharomyces cerevisiae phenotype induced by mitochondrial DNA polymerase mutations associated with progressive external ophthalmoplegia in humans. Hum. Mol. Genet. 15: 2846–2855. - PubMed
-
- Baruffini E., Horvath R., Dallabona C., Czermin B., Lamantea E., et al. , 2011. Predicting the contribution of novel POLG mutations to human disease through analysis in yeast model. Mitochondrion 11: 182–190. - PubMed
-
- Chomyn A., Attardi G., 2003. MtDNA mutations in aging and apoptosis. Biochem. Biophys. Res. Commun. 304: 519–529. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

