A universal RNA structural motif docking the elbow of tRNA in the ribosome, RNAse P and T-box leaders
- PMID: 23580544
- PMCID: PMC3664808
- DOI: 10.1093/nar/gkt219
A universal RNA structural motif docking the elbow of tRNA in the ribosome, RNAse P and T-box leaders
Abstract
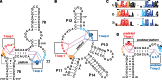
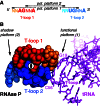
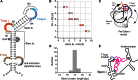
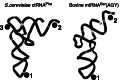
The structure and function of conserved motifs constituting the apex of Stem I in T-box mRNA leaders are investigated. We point out that this apex shares striking similarities with the L1 stalk (helices 76-78) of the ribosome. A sequence and structure analysis of both elements shows that, similarly to the head of the L1 stalk, the function of the apex of Stem I lies in the docking of tRNA through a stacking interaction with the conserved G19:C56 base pair platform. The inferred structure in the apex of Stem I consists of a module of two T-loops bound together head to tail, a module that is also present in the head of the L1 stalk, but went unnoticed. Supporting the analysis, we show that a highly conserved structure in RNAse P formerly described as the J11/12-J12/11 module, which is precisely known to bind the elbow of tRNA, constitutes a third instance of this T-loop module. A structural analysis explains why six nucleotides constituting the core of this module are highly invariant among all three types of RNA. Our finding that major RNA partners of tRNA bind the elbow with a same RNA structure suggests an explanation for the origin of the tRNA L-shape.
Figures


Similar articles
-
Co-crystal structure of a T-box riboswitch stem I domain in complex with its cognate tRNA.Nature. 2013 Aug 15;500(7462):363-6. doi: 10.1038/nature12440. Epub 2013 Jul 28. Nature. 2013. PMID: 23892783 Free PMC article.
-
T box RNA decodes both the information content and geometry of tRNA to affect gene expression.Proc Natl Acad Sci U S A. 2013 Apr 30;110(18):7240-5. doi: 10.1073/pnas.1222214110. Epub 2013 Apr 15. Proc Natl Acad Sci U S A. 2013. PMID: 23589841 Free PMC article.
-
Transfer RNA docking pair model in the ribosomal pre- and post-translocational states.Nucleic Acids Res. 1997 Mar 15;25(6):1254-64. doi: 10.1093/nar/25.6.1254. Nucleic Acids Res. 1997. PMID: 9092637 Free PMC article.
-
Structure of ribonuclease P--a universal ribozyme.Curr Opin Struct Biol. 2006 Jun;16(3):327-35. doi: 10.1016/j.sbi.2006.04.002. Epub 2006 May 2. Curr Opin Struct Biol. 2006. PMID: 16650980 Review.
-
[Formation of spatial structure of RNA molecules].Mol Biol (Mosk). 2012 Jan-Feb;46(1):37-51. Mol Biol (Mosk). 2012. PMID: 22642100 Review. Russian.
Cited by
-
Capture and Release of tRNA by the T-Loop Receptor in the Function of the T-Box Riboswitch.Biochemistry. 2017 Jul 18;56(28):3549-3558. doi: 10.1021/acs.biochem.7b00284. Epub 2017 Jul 3. Biochemistry. 2017. PMID: 28621923 Free PMC article.
-
The tRNA Elbow in Structure, Recognition and Evolution.Life (Basel). 2016 Jan 12;6(1):3. doi: 10.3390/life6010003. Life (Basel). 2016. PMID: 26771646 Free PMC article. Review.
-
Direct observation of tRNA-chaperoned folding of a dynamic mRNA ensemble.Nat Commun. 2023 Sep 6;14(1):5438. doi: 10.1038/s41467-023-41155-3. Nat Commun. 2023. PMID: 37673863 Free PMC article.
-
Molecular envelope and atomic model of an anti-terminated glyQS T-box regulator in complex with tRNAGly.Nucleic Acids Res. 2017 Jul 27;45(13):8079-8090. doi: 10.1093/nar/gkx451. Nucleic Acids Res. 2017. PMID: 28531275 Free PMC article.
-
Structure and mechanism of the T-box riboswitches.Wiley Interdiscip Rev RNA. 2015 Jul-Aug;6(4):419-33. doi: 10.1002/wrna.1285. Epub 2015 May 8. Wiley Interdiscip Rev RNA. 2015. PMID: 25959893 Free PMC article. Review.
References
-
- Grundy FJ, Henkin TM. tRNA as a positive regulator of transcription antitermination in B. subtilis. Cell. 1993;74:475–482. - PubMed
-
- Rollins SM, Grundy FM, Henkin TM. Analysis of cis-acting sequence and structural elements required for antitermination of the Bacillus subtilis tyrS gene. Mol. Microbiol. 1997;25:411–421. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

