Kaposi's sarcoma-associated herpesvirus (KSHV) latency-associated nuclear antigen regulates the KSHV epigenome by association with the histone demethylase KDM3A
- PMID: 23576503
- PMCID: PMC3676133
- DOI: 10.1128/JVI.00011-13
Kaposi's sarcoma-associated herpesvirus (KSHV) latency-associated nuclear antigen regulates the KSHV epigenome by association with the histone demethylase KDM3A
Abstract
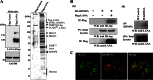
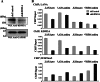
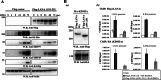
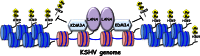
Kaposi's sarcoma-associated herpesvirus (KSHV) latent genomes are tethered to host histones to form a minichromosome also known as an "episome." Histones, which are core components of chromatin, are heavily modified by various histone-targeting enzymes. Posttranslational modifications of histones significantly influence accessibility of transcriptional factors and thus have profound effects on gene expression. Recent studies showed that epigenetic marks on the KSHV episome are well organized, exemplified by the absence of histone H3 lysine 9 (H3K9) methylation, a heterochromatic histone mark, from immediate early and latent gene promoters in naturally infected cells. The present study revealed a mechanistic insight into KSHV epigenome regulation via a complex consisting of LANA and the H3K9me1/2 histone demethylase JMJD1A/KDM3A. This complex was isolated from HeLa cell nuclear extracts stably expressing LANA and was verified by coimmunoprecipitation analyses and with purified proteins. LANA recruitment sites on the KSHV genome inversely correlated with H3K9me2 histone marks in naturally infected cells, and methylation of H3K9 significantly inhibited LANA binding to the histone H3 tail. Chromatin immunoprecipitation coupled with KSHV tiling arrays identified the recruitment sites of the complex, while depletion of LANA expression or overexpression of a KDM3A binding-deficient mutant decreased KDM3A recruitment to the KSHV genome. Finally, ablation of KDM3A expression from latently KSHV-infected cells significantly inhibited KSHV gene expression, leading to decreased KSHV replication during reactivation. Taken together, our results suggest that LANA may play a role in regulation of epigenetic marks on the KSHV genome, which is in part through association with the histone demethylase KDM3A.
Figures




Similar articles
-
Site-specific association with host and viral chromatin by Kaposi's sarcoma-associated herpesvirus LANA and its reversal during lytic reactivation.J Virol. 2014 Jun;88(12):6762-77. doi: 10.1128/JVI.00268-14. Epub 2014 Apr 2. J Virol. 2014. PMID: 24696474 Free PMC article.
-
Kaposi's sarcoma-associated herpesvirus terminal repeat regulates inducible lytic gene promoters.J Virol. 2024 Feb 20;98(2):e0138623. doi: 10.1128/jvi.01386-23. Epub 2024 Jan 19. J Virol. 2024. PMID: 38240593 Free PMC article.
-
The Latency-Associated Nuclear Antigen of Kaposi's Sarcoma-Associated Herpesvirus Inhibits Expression of SUMO/Sentrin-Specific Peptidase 6 To Facilitate Establishment of Latency.J Virol. 2017 Aug 10;91(17):e00806-17. doi: 10.1128/JVI.00806-17. Print 2017 Sep 1. J Virol. 2017. PMID: 28615201 Free PMC article.
-
An atlas of chromatin landscape in KSHV-infected cells during de novo infection and reactivation.Virology. 2024 Sep;597:110146. doi: 10.1016/j.virol.2024.110146. Epub 2024 Jun 19. Virology. 2024. PMID: 38909515 Review.
-
KSHV LANA--the master regulator of KSHV latency.Viruses. 2014 Dec 11;6(12):4961-98. doi: 10.3390/v6124961. Viruses. 2014. PMID: 25514370 Free PMC article. Review.
Cited by
-
A Screen for Extracellular Signal-Regulated Kinase-Primed Glycogen Synthase Kinase 3 Substrates Identifies the p53 Inhibitor iASPP.J Virol. 2015 Sep;89(18):9232-41. doi: 10.1128/JVI.01072-15. Epub 2015 Jun 24. J Virol. 2015. PMID: 26109723 Free PMC article.
-
TRF2 Protein Interacts with Core Histones to Stabilize Chromosome Ends.J Biol Chem. 2016 Sep 23;291(39):20798-810. doi: 10.1074/jbc.M116.719021. Epub 2016 Aug 11. J Biol Chem. 2016. PMID: 27514743 Free PMC article.
-
KSHV vIL-6 enhances inflammatory responses by epigenetic reprogramming.PLoS Pathog. 2023 Nov 7;19(11):e1011771. doi: 10.1371/journal.ppat.1011771. eCollection 2023 Nov. PLoS Pathog. 2023. PMID: 37934757 Free PMC article.
-
MicroRNAs derived from the insect virus HzNV-1 promote lytic infection by suppressing histone methylation.Sci Rep. 2018 Dec 13;8(1):17817. doi: 10.1038/s41598-018-35782-w. Sci Rep. 2018. PMID: 30546025 Free PMC article.
-
Epigenetic control in Kaposi sarcoma-associated herpesvirus infection and associated disease.Semin Immunopathol. 2020 Apr;42(2):143-157. doi: 10.1007/s00281-020-00787-z. Epub 2020 Mar 26. Semin Immunopathol. 2020. PMID: 32219477 Free PMC article. Review.
References
-
- Cesarman E, Chang Y, Moore PS, Said JW, Knowles DM. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N. Engl. J. Med. 332:1186–1191 - PubMed
-
- Dupin N, Fisher C, Kellam P, Ariad S, Tulliez M, Franck N, van Marck E, Salmon D, Gorin I, Escande JP, Weiss RA, Alitalo K, Boshoff C. 1999. Distribution of human herpesvirus-8 latently infected cells in Kaposi's sarcoma, multicentric Castleman's disease, and primary effusion lymphoma. Proc. Natl. Acad. Sci. U. S. A. 96:4546–4551 - PMC - PubMed
-
- Kelley-Clarke B, Ballestas ME, Komatsu T, Kaye KM. 2007. Kaposi's sarcoma herpesvirus C-terminal LANA concentrates at pericentromeric and peri-telomeric regions of a subset of mitotic chromosomes. Virology 357:149–157 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

