Selection of appropriate isolation method based on morphology of blastocyst for efficient derivation of buffalo embryonic stem cells
- PMID: 23553019
- PMCID: PMC3918263
- DOI: 10.1007/s10616-013-9561-7
Selection of appropriate isolation method based on morphology of blastocyst for efficient derivation of buffalo embryonic stem cells
Abstract
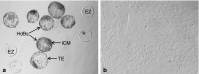
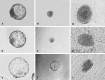
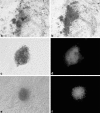
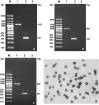
The efficiency of embryonic stem cell (ESC) derivation from all species except for rodents and primates is very low. There are however, multiple interests in obtaining pluripotent cells from these animals with main expectations in the fields of transgenesis, cloning, regenerative medicine and tissue engineering. Researches are being carried out in laboratories throughout the world to increase the efficiency of ESC isolation for their downstream applications. Thus, the present study was undertaken to study the effect of different isolation methods based on the morphology of blastocyst for efficient derivation of buffalo ESCs. Embryos were produced in vitro through the procedures of maturation, fertilization and culture. Hatched blastocysts or isolated inner cell masses (ICMs) were seeded on mitomycin-C inactivated buffalo fetal fibroblast monolayer for the development of ESC colonies. The ESCs were analyzed for alkaline phosphatase activity, expression of pluripotency markers and karyotypic stability. Primary ESC colonies were obtained after 2-5 days of seeding hatched blastocysts or isolated ICMs on mitomycin-C inactivated feeder layer. Mechanically isolated ICMs attached and formed primary cell colonies more efficiently than ICMs isolated enzymatically. For derivation of ESCs from poorly defined ICMs intact hatched blastocyst culture was the most successful method. Results of this study implied that although ESCs can be obtained using all three methods used in this study, efficiency varies depending upon the morphology of blastocyst and isolation method used. So, appropriate isolation method must be selected depending on the quality of blastocyst for efficient derivation of ESCs.
Figures
Similar articles
-
Expression and quantification of Oct-4 gene in blastocyst and embryonic stem cells derived from in vitro produced buffalo embryos.In Vitro Cell Dev Biol Anim. 2012 Apr;48(4):229-35. doi: 10.1007/s11626-012-9491-2. Epub 2012 Mar 22. In Vitro Cell Dev Biol Anim. 2012. PMID: 22438133
-
In vivo differentiation potential of buffalo (Bubalus bubalis) embryonic stem cell.In Vitro Cell Dev Biol Anim. 2012 Jun;48(6):349-58. doi: 10.1007/s11626-012-9515-y. Epub 2012 Jun 8. In Vitro Cell Dev Biol Anim. 2012. PMID: 22678753
-
Derivation of buffalo embryonic stem-like cells from in vitro-produced blastocysts on homologous and heterologous feeder cells.J Assist Reprod Genet. 2011 Aug;28(8):679-88. doi: 10.1007/s10815-011-9572-2. Epub 2011 May 4. J Assist Reprod Genet. 2011. PMID: 21573679 Free PMC article.
-
Nonviable human pre-implantation embryos as a source of stem cells for research and potential therapy.Stem Cell Rev. 2005 Dec;1(4):337-43. doi: 10.1385/SCR:1:4:337. Stem Cell Rev. 2005. PMID: 17142877 Review.
-
Searching for naïve human pluripotent stem cells.World J Stem Cells. 2015 Apr 26;7(3):649-56. doi: 10.4252/wjsc.v7.i3.649. World J Stem Cells. 2015. PMID: 25914771 Free PMC article. Review.
References
-
- Anand T, Kumar D, Singh MK, Shah RA, Chauhan MS, Manik RS, Singla SK, Palta P. Buffalo (Bubalus bubalis) embryonic stem cell-like cells and preimplantation embryos exhibit comparable expression of pluripotency-related antigens. Reprod Domest Anim. 2011;46:50–58. doi: 10.1111/j.1439-0531.2009.01564.x. - DOI - PubMed
-
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, Gifford DK, Melton DA, Jaenisch R, Young RA. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

