Genome-scale analyses of butanol tolerance in Saccharomyces cerevisiae reveal an essential role of protein degradation
- PMID: 23552365
- PMCID: PMC3621596
- DOI: 10.1186/1754-6834-6-48
Genome-scale analyses of butanol tolerance in Saccharomyces cerevisiae reveal an essential role of protein degradation
Abstract
Background: n-Butanol and isobutanol produced from biomass-derived sugars are promising renewable transport fuels and solvents. Saccharomyces cerevisiae has been engineered for butanol production, but its high butanol sensitivity poses an upper limit to product titers that can be reached by further pathway engineering. A better understanding of the molecular basis of butanol stress and tolerance of S. cerevisiae is important for achieving improved tolerance.
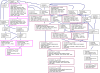
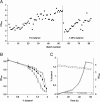
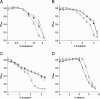
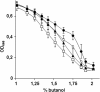
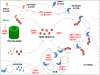
Results: By combining a screening of the haploid S. cerevisiae knock-out library, gene overexpression, and genome analysis of evolutionary engineered n-butanol-tolerant strains, we established that protein degradation plays an essential role in tolerance. Strains deleted in genes involved in the ubiquitin-proteasome system and in vacuolar degradation of damaged proteins showed hypersensitivity to n-butanol. Overexpression of YLR224W, encoding the subunit responsible for the recognition of damaged proteins of an ubiquitin ligase complex, resulted in a strain with a higher n-butanol tolerance. Two independently evolved n-butanol-tolerant strains carried different mutations in both RPN4 and RTG1, which encode transcription factors involved in the expression of proteasome and peroxisomal genes, respectively. Introduction of these mutated alleles in the reference strain increased butanol tolerance, confirming their relevance in the higher tolerance phenotype. The evolved strains, in addition to n-butanol, were also more tolerant to 2-butanol, isobutanol and 1-propanol, indicating a common molecular basis for sensitivity and tolerance to C3 and C4 alcohols.
Conclusions: This study shows that maintenance of protein integrity plays an essential role in butanol tolerance and demonstrates new promising targets to engineer S. cerevisiae for improved tolerance.
Figures

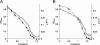

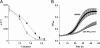

Similar articles
-
Phenotypic characterisation of Saccharomyces spp. for tolerance to 1-butanol.J Ind Microbiol Biotechnol. 2014 Nov;41(11):1627-36. doi: 10.1007/s10295-014-1511-7. Epub 2014 Sep 23. J Ind Microbiol Biotechnol. 2014. PMID: 25242291
-
Physiological adaptations of Saccharomyces cerevisiae evolved for improved butanol tolerance.Biotechnol Biofuels. 2013 Jul 15;6(1):101. doi: 10.1186/1754-6834-6-101. Biotechnol Biofuels. 2013. PMID: 23855998 Free PMC article.
-
Evolutionary engineering improves tolerance for medium-chain alcohols in Saccharomyces cerevisiae.Biotechnol Biofuels. 2018 Apr 2;11:90. doi: 10.1186/s13068-018-1089-9. eCollection 2018. Biotechnol Biofuels. 2018. PMID: 29619086 Free PMC article.
-
Butanol production by Saccharomyces cerevisiae: perspectives, strategies and challenges.World J Microbiol Biotechnol. 2020 Mar 9;36(3):48. doi: 10.1007/s11274-020-02828-z. World J Microbiol Biotechnol. 2020. PMID: 32152786 Review.
-
Metabolic engineering of Saccharomyces cerevisiae for production of butanol isomers.Curr Opin Biotechnol. 2015 Jun;33:1-7. doi: 10.1016/j.copbio.2014.09.004. Epub 2014 Oct 4. Curr Opin Biotechnol. 2015. PMID: 25286420 Review.
Cited by
-
Rpn4 and proteasome-mediated yeast resistance to ethanol includes regulation of autophagy.Appl Microbiol Biotechnol. 2020 May;104(9):4027-4041. doi: 10.1007/s00253-020-10518-x. Epub 2020 Mar 10. Appl Microbiol Biotechnol. 2020. PMID: 32157425
-
Genome replication engineering assisted continuous evolution (GREACE) to improve microbial tolerance for biofuels production.Biotechnol Biofuels. 2013 Sep 27;6(1):137. doi: 10.1186/1754-6834-6-137. Biotechnol Biofuels. 2013. PMID: 24070173 Free PMC article.
-
Functional toxicology: tools to advance the future of toxicity testing.Front Genet. 2014 May 5;5:110. doi: 10.3389/fgene.2014.00110. eCollection 2014. Front Genet. 2014. PMID: 24847352 Free PMC article. Review.
-
Alternative reactions at the interface of glycolysis and citric acid cycle in Saccharomyces cerevisiae.FEMS Yeast Res. 2016 May;16(3):fow017. doi: 10.1093/femsyr/fow017. Epub 2016 Feb 18. FEMS Yeast Res. 2016. PMID: 26895788 Free PMC article.
-
ChiNet uncovers rewired transcription subnetworks in tolerant yeast for advanced biofuels conversion.Nucleic Acids Res. 2015 May 19;43(9):4393-407. doi: 10.1093/nar/gkv358. Epub 2015 Apr 20. Nucleic Acids Res. 2015. PMID: 25897127 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

