Investigation of radiation-induced transcriptome profile of radioresistant non-small cell lung cancer A549 cells using RNA-seq
- PMID: 23533613
- PMCID: PMC3606344
- DOI: 10.1371/journal.pone.0059319
Investigation of radiation-induced transcriptome profile of radioresistant non-small cell lung cancer A549 cells using RNA-seq
Abstract
Radioresistance is a main impediment to effective radiotherapy for non-small cell lung cancer (NSCLC). Despite several experimental and clinical studies of resistance to radiation, the precise mechanism of radioresistance in NSCLC cells and tissues still remains unclear. This result could be explained by limitation of previous researches such as a partial understanding of the cellular radioresistance mechanism at a single molecule level. In this study, we aimed to investigate extensive radiation responses in radioresistant NSCLC cells and to identify radioresistance-associating factors. For the first time, using RNA-seq, a massive sequencing-based approach, we examined whole-transcriptome alteration in radioresistant NSCLC A549 cells under irradiation, and verified significant radiation-altered genes and their chromosome distribution patterns. Also, bioinformatic approaches (GO analysis and IPA) were performed to characterize the radiation responses in radioresistant A549 cells. We found that epithelial-mesenchymal transition (EMT), migration and inflammatory processes could be meaningfully related to regulation of radiation responses in radioresistant A549 cells. Based on the results of bioinformatic analysis for the radiation-induced transcriptome alteration, we selected seven significant radiation-altered genes (SESN2, FN1, TRAF4, CDKN1A, COX-2, DDB2 and FDXR) and then compared radiation effects in two types of NSCLC cells with different radiosensitivity (radioresistant A549 cells and radiosensitive NCI-H460 cells). Interestingly, under irradiation, COX-2 showed the most significant difference in mRNA and protein expression between A549 and NCI-H460 cells. IR-induced increase of COX-2 expression was appeared only in radioresistant A549 cells. Collectively, we suggest that COX-2 (also known as prostaglandin-endoperoxide synthase 2 (PTGS2)) could have possibility as a putative biomarker for radioresistance in NSCLC cells.
Conflict of interest statement
Figures

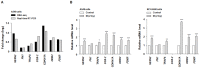

Similar articles
-
ITGB1 enhances the Radioresistance of human Non-small Cell Lung Cancer Cells by modulating the DNA damage response and YAP1-induced Epithelial-mesenchymal Transition.Int J Biol Sci. 2021 Jan 18;17(2):635-650. doi: 10.7150/ijbs.52319. eCollection 2021. Int J Biol Sci. 2021. PMID: 33613118 Free PMC article.
-
miR-219a-5p enhances the radiosensitivity of non-small cell lung cancer cells through targeting CD164.Biosci Rep. 2020 Jul 31;40(7):BSR20192795. doi: 10.1042/BSR20192795. Biosci Rep. 2020. PMID: 32364222 Free PMC article.
-
Transcriptome-Based Traits of Radioresistant Sublines of Non-Small Cell Lung Cancer Cells.Int J Mol Sci. 2023 Feb 3;24(3):3042. doi: 10.3390/ijms24033042. Int J Mol Sci. 2023. PMID: 36769365 Free PMC article.
-
Attenuated LKB1-SIK1 signaling promotes epithelial-mesenchymal transition and radioresistance of non-small cell lung cancer cells.Chin J Cancer. 2016 Jun 7;35:50. doi: 10.1186/s40880-016-0113-3. Chin J Cancer. 2016. PMID: 27266881 Free PMC article.
-
CLPTM1L induces estrogen receptor β signaling-mediated radioresistance in non-small cell lung cancer cells.Cell Commun Signal. 2020 Sep 17;18(1):152. doi: 10.1186/s12964-020-00571-4. Cell Commun Signal. 2020. PMID: 32943060 Free PMC article.
Cited by
-
TGF-β receptor inhibitor LY2109761 enhances the radiosensitivity of gastric cancer by inactivating the TGF-β/SMAD4 signaling pathway.Aging (Albany NY). 2019 Oct 19;11(20):8892-8910. doi: 10.18632/aging.102329. Epub 2019 Oct 19. Aging (Albany NY). 2019. PMID: 31631064 Free PMC article.
-
TRIAP1 knockdown sensitizes non-small cell lung cancer to ionizing radiation by disrupting redox homeostasis.Thorac Cancer. 2020 Apr;11(4):1015-1025. doi: 10.1111/1759-7714.13358. Epub 2020 Feb 25. Thorac Cancer. 2020. PMID: 32096592 Free PMC article.
-
Global profiling of transcriptome, proteome and 2-hydroxyisobutyrylome in radioresistant lung adenocarcinoma cell.BMC Genomics. 2024 Oct 3;25(1):923. doi: 10.1186/s12864-024-10854-6. BMC Genomics. 2024. PMID: 39363283 Free PMC article.
-
Overexpression of DUSP6 enhances chemotherapy-resistance of ovarian epithelial cancer by regulating the ERK signaling pathway.J Cancer. 2020 Mar 4;11(11):3151-3164. doi: 10.7150/jca.37267. eCollection 2020. J Cancer. 2020. PMID: 32231719 Free PMC article.
-
Inhibition of DNA‑PK activity sensitizes A549 cells to X‑ray irradiation by inducing the ATM‑dependent DNA damage response.Mol Med Rep. 2018 Jun;17(6):7545-7552. doi: 10.3892/mmr.2018.8828. Epub 2018 Mar 29. Mol Med Rep. 2018. PMID: 29620203 Free PMC article.
References
-
- Yin ZJ, Jin FG, Liu TG, Fu EQ, Xie YH, et al. (2011) Overexpression of STAT3 potentiates growth, survival, and radioresistance of non-small-cell lung cancer (NSCLC) cells. J Surg Res 171: 675–683. - PubMed
-
- Shin S, Cha HJ, Lee EM, Lee SJ, Seo SK, et al. (2009) Alteration of miRNA profiles by ionizing radiation in A549 human non-small cell lung cancer cells. Int J Oncol 35: 81–86. - PubMed
-
- Kim W, Youn H, Seong KM, Yang HJ, Yun YJ, et al. (2011) PIM1-activated PRAS40 regulates radioresistance in non-small cell lung cancer cells through interplay with FOXO3a, 14-3-3 and protein phosphatases. Radiat Res 176: 539–552. - PubMed
-
- Liang K, Ang KK, Milas L, Hunter N, Fan Z (2003) The epidermal growth factor receptor mediates radioresistance. Int J Radiat Oncol Biol Phys 57: 246–254. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

