A separate pool of cardiac phospholemman that does not regulate or associate with the sodium pump: multimers of phospholemman in ventricular muscle
- PMID: 23532852
- PMCID: PMC3650417
- DOI: 10.1074/jbc.M113.460956
A separate pool of cardiac phospholemman that does not regulate or associate with the sodium pump: multimers of phospholemman in ventricular muscle
Abstract
Background: Phospholemman regulates the plasmalemmal sodium pump in excitable tissues.
Results: In cardiac muscle, a subpopulation of phospholemman with a unique phosphorylation signature associates with other phospholemman molecules but not with the pump.
Conclusion: Phospholemman oligomers exist in cardiac muscle.
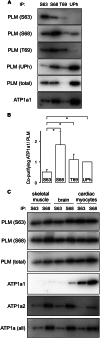
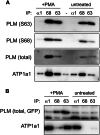
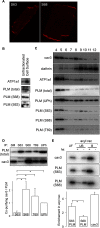
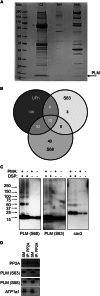
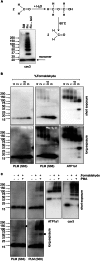
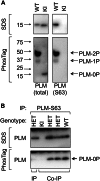
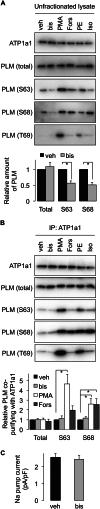
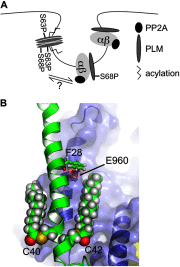
Significance: Much like phospholamban regulation of SERCA, phospholemman exists as both a sodium pump inhibiting monomer and an unassociated oligomer. Phospholemman (PLM), the principal quantitative sarcolemmal substrate for protein kinases A and C in the heart, regulates the cardiac sodium pump. Much like phospholamban, which regulates the related ATPase SERCA, PLM is reported to oligomerize. We investigated subpopulations of PLM in adult rat ventricular myocytes based on phosphorylation status. Co-immunoprecipitation identified two pools of PLM: one not associated with the sodium pump phosphorylated at Ser(63) and one associated with the pump, both phosphorylated at Ser(68) and unphosphorylated. Phosphorylation of PLM at Ser(63) following activation of PKC did not abrogate association of PLM with the pump, so its failure to associate with the pump was not due to phosphorylation at this site. All pools of PLM co-localized to cell surface caveolin-enriched microdomains with sodium pump α subunits, despite the lack of caveolin-binding motif in PLM. Mass spectrometry analysis of phosphospecific immunoprecipitation reactions revealed no unique protein interactions for Ser(63)-phosphorylated PLM, and cross-linking reagents also failed to identify any partner proteins for this pool. In lysates from hearts of heterozygous transgenic animals expressing wild type and unphosphorylatable PLM, Ser(63)-phosphorylated PLM co-immunoprecipitated unphosphorylatable PLM, confirming the existence of PLM multimers. Dephosphorylation of the PLM multimer does not change sodium pump activity. Hence like phospholamban, PLM exists as a pump-inhibiting monomer and an unassociated oligomer. The distribution of different PLM phosphorylation states to different pools may be explained by their differential proximity to protein phosphatases rather than a direct effect of phosphorylation on PLM association with the pump.
Keywords: Caveolae; FXYD Proteins; Heart; Na,K-ATPase; PP2A; Phospholemman; Protein Palmitoylation; Protein Phosphatase; Protein Phosphorylation; Serine-Threonine Protein Phosphatase.
Figures
Similar articles
-
The inhibitory effect of phospholemman on the sodium pump requires its palmitoylation.J Biol Chem. 2011 Oct 14;286(41):36020-36031. doi: 10.1074/jbc.M111.282145. Epub 2011 Aug 25. J Biol Chem. 2011. PMID: 21868384 Free PMC article.
-
Nitric oxide regulates cardiac intracellular Na⁺ and Ca²⁺ by modulating Na/K ATPase via PKCε and phospholemman-dependent mechanism.J Mol Cell Cardiol. 2013 Aug;61:164-71. doi: 10.1016/j.yjmcc.2013.04.013. Epub 2013 Apr 20. J Mol Cell Cardiol. 2013. PMID: 23612119 Free PMC article.
-
Protein Phosphatase 1c Associated with the Cardiac Sodium Calcium Exchanger 1 Regulates Its Activity by Dephosphorylating Serine 68-phosphorylated Phospholemman.J Biol Chem. 2016 Feb 26;291(9):4561-79. doi: 10.1074/jbc.M115.677898. Epub 2015 Dec 14. J Biol Chem. 2016. PMID: 26668322 Free PMC article.
-
Regulation of the cardiac sodium pump.Cell Mol Life Sci. 2013 Apr;70(8):1357-80. doi: 10.1007/s00018-012-1134-y. Epub 2012 Sep 7. Cell Mol Life Sci. 2013. PMID: 22955490 Free PMC article. Review.
-
Coordinated regulation of cardiac Na(+)/Ca (2+) exchanger and Na (+)-K (+)-ATPase by phospholemman (FXYD1).Adv Exp Med Biol. 2013;961:175-90. doi: 10.1007/978-1-4614-4756-6_15. Adv Exp Med Biol. 2013. PMID: 23224879 Review.
Cited by
-
Phospholemman is not required for the acute stimulation of Na⁺-K⁺-ATPase α₂-activity during skeletal muscle fatigue.Am J Physiol Cell Physiol. 2015 Dec 15;309(12):C813-22. doi: 10.1152/ajpcell.00205.2015. Epub 2015 Oct 14. Am J Physiol Cell Physiol. 2015. PMID: 26468207 Free PMC article.
-
Phospholemman Phosphorylation Regulates Vascular Tone, Blood Pressure, and Hypertension in Mice and Humans.Circulation. 2021 Mar 16;143(11):1123-1138. doi: 10.1161/CIRCULATIONAHA.119.040557. Epub 2020 Dec 18. Circulation. 2021. PMID: 33334125 Free PMC article.
-
Structure of the Na,K-ATPase regulatory protein FXYD2b in micelles: implications for membrane-water interfacial arginines.Biochim Biophys Acta. 2015 Jan;1848(1 Pt B):299-306. doi: 10.1016/j.bbamem.2014.04.021. Epub 2014 May 2. Biochim Biophys Acta. 2015. PMID: 24794573 Free PMC article.
-
BAG3 regulates contractility and Ca(2+) homeostasis in adult mouse ventricular myocytes.J Mol Cell Cardiol. 2016 Mar;92:10-20. doi: 10.1016/j.yjmcc.2016.01.015. Epub 2016 Jan 19. J Mol Cell Cardiol. 2016. PMID: 26796036 Free PMC article.
-
Sarcolipin alters SERCA1a interdomain communication by impairing binding of both calcium and ATP.Sci Rep. 2021 Jan 15;11(1):1641. doi: 10.1038/s41598-021-81061-6. Sci Rep. 2021. PMID: 33452371 Free PMC article.
References
-
- Sweadner K. J., Rael E. (2000) The FXYD gene family of small ion transport regulators or channels. cDNA sequence, protein signature sequence, and expression. Genomics 68, 41–56 - PubMed
-
- Geering K. (2006) FXYD proteins. New regulators of Na-K-ATPase. Am. J. Physiol. Renal Physiol. 290, F241–F250 - PubMed
-
- Fuller W., Eaton P., Bell J. R., Shattock M. J. (2004) Ischemia-induced phosphorylation of phospholemman directly activates rat cardiac Na/K-ATPase. FASEB J. 18, 197–199 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

