Redistribution of H3K27me3 upon DNA hypomethylation results in de-repression of Polycomb target genes
- PMID: 23531360
- PMCID: PMC4053768
- DOI: 10.1186/gb-2013-14-3-r25
Redistribution of H3K27me3 upon DNA hypomethylation results in de-repression of Polycomb target genes
Abstract
Background: DNA methylation and the Polycomb repression system are epigenetic mechanisms that play important roles in maintaining transcriptional repression. Recent evidence suggests that DNA methylation can attenuate the binding of Polycomb protein components to chromatin and thus plays a role in determining their genomic targeting. However, whether this role of DNA methylation is important in the context of transcriptional regulation is unclear.
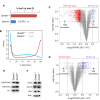
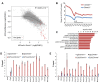
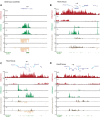
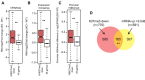
Results: By genome-wide mapping of the Polycomb Repressive Complex 2-signature histone mark, H3K27me3, in severely DNA hypomethylated mouse somatic cells, we show that hypomethylation leads to widespread H3K27me3 redistribution, in a manner that reflects the local DNA methylation status in wild-type cells. Unexpectedly, we observe striking loss of H3K27me3 and Polycomb Repressive Complex 2 from Polycomb target gene promoters in DNA hypomethylated cells, including Hox gene clusters. Importantly, we show that many of these genes become ectopically expressed in DNA hypomethylated cells, consistent with loss of Polycomb-mediated repression.
Conclusions: An intact DNA methylome is required for appropriate Polycomb-mediated gene repression by constraining Polycomb Repressive Complex 2 targeting. These observations identify a previously unappreciated role for DNA methylation in gene regulation and therefore influence our understanding of how this epigenetic mechanism contributes to normal development and disease.
Figures


Similar articles
-
Targeting of EZH2 to a defined genomic site is sufficient for recruitment of Dnmt3a but not de novo DNA methylation.Epigenetics. 2009 Aug 16;4(6):404-14. doi: 10.4161/epi.4.6.9392. Epub 2009 Aug 29. Epigenetics. 2009. PMID: 19717977
-
Signatures of polycomb repression and reduced H3K4 trimethylation are associated with p15INK4b DNA methylation in AML.Blood. 2010 Apr 15;115(15):3098-108. doi: 10.1182/blood-2009-07-233858. Epub 2010 Feb 26. Blood. 2010. PMID: 20190193 Free PMC article.
-
Cooperation between the H3K27me3 Chromatin Mark and Non-CG Methylation in Epigenetic Regulation.Plant Physiol. 2016 Oct;172(2):1131-1141. doi: 10.1104/pp.16.01238. Epub 2016 Aug 17. Plant Physiol. 2016. PMID: 27535791 Free PMC article.
-
Not just a writer: PRC2 as a chromatin reader.Biochem Soc Trans. 2021 Jun 30;49(3):1159-1170. doi: 10.1042/BST20200728. Biochem Soc Trans. 2021. PMID: 34060617 Free PMC article. Review.
-
EZH2 methyltransferase and H3K27 methylation in breast cancer.Int J Biol Sci. 2012;8(1):59-65. doi: 10.7150/ijbs.8.59. Epub 2011 Nov 18. Int J Biol Sci. 2012. PMID: 22211105 Free PMC article. Review.
Cited by
-
The renaissance and enlightenment of Marchantia as a model system.Plant Cell. 2022 Sep 27;34(10):3512-3542. doi: 10.1093/plcell/koac219. Plant Cell. 2022. PMID: 35976122 Free PMC article.
-
Dihydroartemisinin inhibits prostate cancer via JARID2/miR-7/miR-34a-dependent downregulation of Axl.Oncogenesis. 2019 Feb 19;8(3):14. doi: 10.1038/s41389-019-0122-6. Oncogenesis. 2019. PMID: 30783079 Free PMC article.
-
Redistribution of H3K27me3 and acetylated histone H4 upon exposure to azacitidine and decitabine results in de-repression of the AML1/ETO target gene IL3.Epigenetics. 2014 Mar;9(3):387-95. doi: 10.4161/epi.27322. Epub 2013 Dec 2. Epigenetics. 2014. PMID: 24300456 Free PMC article.
-
Telomere dysfunction cooperates with epigenetic alterations to impair murine embryonic stem cell fate commitment.Elife. 2020 Apr 16;9:e47333. doi: 10.7554/eLife.47333. Elife. 2020. PMID: 32297856 Free PMC article.
-
The quest for mammalian Polycomb response elements: are we there yet?Chromosoma. 2016 Jun;125(3):471-96. doi: 10.1007/s00412-015-0539-4. Epub 2015 Oct 9. Chromosoma. 2016. PMID: 26453572 Free PMC article. Review.
References
-
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, Edsall L, Antosiewicz-Bourget J, Stewart R, Ruotti V, Millar AH, Thomson JA, Ren B, Ecker JR. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;14:315–322. doi: 10.1038/nature08514. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

