Glutamate receptor desensitization is mediated by changes in quaternary structure of the ligand binding domain
- PMID: 23530186
- PMCID: PMC3625259
- DOI: 10.1073/pnas.1217549110
Glutamate receptor desensitization is mediated by changes in quaternary structure of the ligand binding domain
Abstract
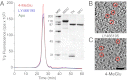
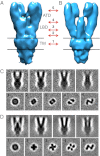
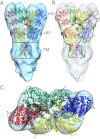
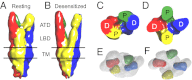
Glutamate receptor ion channels are membrane proteins that mediate excitatory synaptic transmission in the central nervous system of vertebrates. Insight into molecular mechanisms underlying glutamate receptor gating is limited by lack of structural information for receptors trapped in different conformational states. Here, we report the use of single-particle cryoelectron tomography to determine the structures, at ∼21 Å resolution, of full-length GluK2 kainate receptors trapped in antagonist-bound resting and agonist-bound desensitized states. The resting state, stabilized by the competitive antagonist LY466195, closely resembles the crystal structure of the AMPA receptor GluA2, with well-resolved proximal and distal subunits exhibiting cross-over between the twofold symmetric amino terminal domain and a twofold symmetric ligand binding domain (LBD) dimer of dimers assembly. In the desensitized state, the LBD undergoes a major rearrangement, resulting in a separation of the four subunits by ∼25 Å. However, the amino terminal domain, transmembrane, and cytoplasmic regions of the receptor have similar conformations in the resting and desensitized states. The LBD rearrangement was not anticipated in prior models based on crystal structures for soluble LBD dimer assemblies, and we speculate that subunit separation allows a better match to the fourfold symmetric ion channel domain. From fits of the amino terminal domain and LBD domains into the density map of the desensitized state we have derived a structural model for differences in quaternary conformation between the resting and desensitized states.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Structure and symmetry inform gating principles of ionotropic glutamate receptors.Neuropharmacology. 2017 Jan;112(Pt A):11-15. doi: 10.1016/j.neuropharm.2016.08.034. Epub 2016 Sep 20. Neuropharmacology. 2017. PMID: 27663701 Free PMC article. Review.
-
Domain organization and function in GluK2 subtype kainate receptors.Proc Natl Acad Sci U S A. 2010 May 4;107(18):8463-8. doi: 10.1073/pnas.1000838107. Epub 2010 Apr 19. Proc Natl Acad Sci U S A. 2010. PMID: 20404149 Free PMC article.
-
Structural basis of kainate subtype glutamate receptor desensitization.Nature. 2016 Sep 22;537(7621):567-571. doi: 10.1038/nature19352. Epub 2016 Aug 31. Nature. 2016. PMID: 27580033 Free PMC article.
-
Structural mechanism of glutamate receptor activation and desensitization.Nature. 2014 Oct 16;514(7522):328-34. doi: 10.1038/nature13603. Epub 2014 Aug 3. Nature. 2014. PMID: 25119039 Free PMC article.
-
Lessons from crystal structures of kainate receptors.Neuropharmacology. 2017 Jan;112(Pt A):16-28. doi: 10.1016/j.neuropharm.2016.05.014. Epub 2016 May 26. Neuropharmacology. 2017. PMID: 27236079 Review.
Cited by
-
Structure and symmetry inform gating principles of ionotropic glutamate receptors.Neuropharmacology. 2017 Jan;112(Pt A):11-15. doi: 10.1016/j.neuropharm.2016.08.034. Epub 2016 Sep 20. Neuropharmacology. 2017. PMID: 27663701 Free PMC article. Review.
-
The N-terminal domain modulates α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor desensitization.J Biol Chem. 2014 May 9;289(19):13197-205. doi: 10.1074/jbc.M113.526301. Epub 2014 Mar 20. J Biol Chem. 2014. PMID: 24652293 Free PMC article.
-
Structural Arrangement Produced by Concanavalin A Binding to Homomeric GluK2 Receptors.Membranes (Basel). 2021 Aug 11;11(8):613. doi: 10.3390/membranes11080613. Membranes (Basel). 2021. PMID: 34436376 Free PMC article.
-
Protein Crowding within the Postsynaptic Density Can Impede the Escape of Membrane Proteins.J Neurosci. 2016 Apr 13;36(15):4276-95. doi: 10.1523/JNEUROSCI.3154-15.2016. J Neurosci. 2016. PMID: 27076425 Free PMC article.
-
Glycine activated ion channel subunits encoded by ctenophore glutamate receptor genes.Proc Natl Acad Sci U S A. 2015 Nov 3;112(44):E6048-57. doi: 10.1073/pnas.1513771112. Epub 2015 Oct 12. Proc Natl Acad Sci U S A. 2015. PMID: 26460032 Free PMC article.
References
-
- Shepherd JD, Huganir RL. The cell biology of synaptic plasticity: AMPA receptor trafficking. Annu Rev Cell Dev Biol. 2007;23:613–643. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

