Characterization of transcriptional networks in blood stem and progenitor cells using high-throughput single-cell gene expression analysis
- PMID: 23524953
- PMCID: PMC3796878
- DOI: 10.1038/ncb2709
Characterization of transcriptional networks in blood stem and progenitor cells using high-throughput single-cell gene expression analysis
Erratum in
- Nat Cell Biol. 2013 May;15(5):544
Abstract
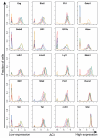
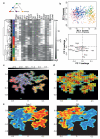
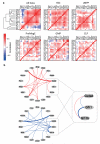
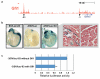
Cellular decision-making is mediated by a complex interplay of external stimuli with the intracellular environment, in particular transcription factor regulatory networks. Here we have determined the expression of a network of 18 key haematopoietic transcription factors in 597 single primary blood stem and progenitor cells isolated from mouse bone marrow. We demonstrate that different stem/progenitor populations are characterized by distinctive transcription factor expression states, and through comprehensive bioinformatic analysis reveal positively and negatively correlated transcription factor pairings, including previously unrecognized relationships between Gata2, Gfi1 and Gfi1b. Validation using transcriptional and transgenic assays confirmed direct regulatory interactions consistent with a regulatory triad in immature blood stem cells, where Gata2 may function to modulate cross-inhibition between Gfi1 and Gfi1b. Single-cell expression profiling therefore identifies network states and allows reconstruction of network hierarchies involved in controlling stem cell fate choices, and provides a blueprint for studying both normal development and human disease.
Figures

Similar articles
-
C/EBPa controls acquisition and maintenance of adult haematopoietic stem cell quiescence.Nat Cell Biol. 2013 Apr;15(4):385-94. doi: 10.1038/ncb2698. Epub 2013 Mar 17. Nat Cell Biol. 2013. PMID: 23502316 Free PMC article.
-
Nrf2 regulates haematopoietic stem cell function.Nat Cell Biol. 2013 Mar;15(3):309-16. doi: 10.1038/ncb2699. Epub 2013 Feb 24. Nat Cell Biol. 2013. PMID: 23434824 Free PMC article.
-
A network including TGFβ/Smad4, Gata2, and p57 regulates proliferation of mouse hematopoietic progenitor cells.Exp Hematol. 2016 May;44(5):399-409.e5. doi: 10.1016/j.exphem.2016.02.001. Epub 2016 Feb 10. Exp Hematol. 2016. PMID: 26876150
-
Advancing haematopoietic stem and progenitor cell biology through single-cell profiling.FEBS Lett. 2016 Nov;590(22):4052-4067. doi: 10.1002/1873-3468.12231. Epub 2016 Jun 21. FEBS Lett. 2016. PMID: 27259698 Review.
-
From cytopenia to leukemia: the role of Gfi1 and Gfi1b in blood formation.Blood. 2015 Dec 10;126(24):2561-9. doi: 10.1182/blood-2015-06-655043. Epub 2015 Oct 7. Blood. 2015. PMID: 26447191 Free PMC article. Review.
Cited by
-
3D computational reconstruction of tissues with hollow spherical morphologies using single-cell gene expression data.Nat Protoc. 2015 Mar;10(3):459-474. doi: 10.1038/nprot.2015.022. Epub 2015 Feb 12. Nat Protoc. 2015. PMID: 25675210 Free PMC article.
-
Hematopoiesis and T-cell specification as a model developmental system.Immunol Rev. 2016 May;271(1):72-97. doi: 10.1111/imr.12417. Immunol Rev. 2016. PMID: 27088908 Free PMC article. Review.
-
Data-driven modeling predicts gene regulatory network dynamics during the differentiation of multipotential hematopoietic progenitors.PLoS Comput Biol. 2022 Jan 14;18(1):e1009779. doi: 10.1371/journal.pcbi.1009779. eCollection 2022 Jan. PLoS Comput Biol. 2022. PMID: 35030198 Free PMC article.
-
Single-cell transcriptional uncertainty landscape of cell differentiation.F1000Res. 2023 Jul 20;12:426. doi: 10.12688/f1000research.131861.2. eCollection 2023. F1000Res. 2023. PMID: 37545651 Free PMC article.
-
Executable cancer models: successes and challenges.Nat Rev Cancer. 2020 Jun;20(6):343-354. doi: 10.1038/s41568-020-0258-x. Epub 2020 Apr 27. Nat Rev Cancer. 2020. PMID: 32341552 Review.
References
-
- Ottersbach K, Smith A, Wood A, Gottgens B. Ontogeny of haematopoiesis: recent advances and open questions. Br J Haematol. 2010;148:343–355. - PubMed
-
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. - PubMed
-
- Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. - PubMed
-
- Gering M, Yamada Y, Rabbitts TH, Patient RK. Lmo2 and Scl/Tal1 convert non-axial mesoderm into haemangioblasts which differentiate into endothelial cells in the absence of Gata1. Development. 2003;130:6187–6199. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

