Discovering host genes involved in the infection by the Tomato Yellow Leaf Curl Virus complex and in the establishment of resistance to the virus using Tobacco Rattle Virus-based post transcriptional gene silencing
- PMID: 23524390
- PMCID: PMC3705308
- DOI: 10.3390/v5030998
Discovering host genes involved in the infection by the Tomato Yellow Leaf Curl Virus complex and in the establishment of resistance to the virus using Tobacco Rattle Virus-based post transcriptional gene silencing
Abstract
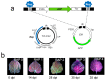
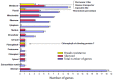
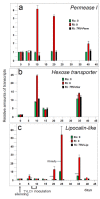
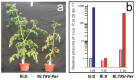
The development of high-throughput technologies allows for evaluating gene expression at the whole-genome level. Together with proteomic and metabolomic studies, these analyses have resulted in the identification of plant genes whose function or expression is altered as a consequence of pathogen attacks. Members of the Tomato yellow leaf curl virus (TYLCV) complex are among the most important pathogens impairing production of agricultural crops worldwide. To understand how these geminiviruses subjugate plant defenses, and to devise counter-measures, it is essential to identify the host genes affected by infection and to determine their role in susceptible and resistant plants. We have used a reverse genetics approach based on Tobacco rattle virus-induced gene silencing (TRV-VIGS) to uncover genes involved in viral infection of susceptible plants, and to identify genes underlying virus resistance. To identify host genes with a role in geminivirus infection, we have engineered a Nicotiana benthamiana line, coined 2IRGFP, which over-expresses GFP upon virus infection. With this system, we have achieved an accurate description of the dynamics of virus replication in space and time. Upon silencing selected N. benthamiana genes previously shown to be related to host response to geminivirus infection, we have identified eighteen genes involved in a wide array of cellular processes. Plant genes involved in geminivirus resistance were studied by comparing two tomato lines: one resistant (R), the other susceptible (S) to the virus. Sixty-nine genes preferentially expressed in R tomatoes were identified by screening cDNA libraries from infected and uninfected R and S genotypes. Out of the 25 genes studied so far, the silencing of five led to the total collapse of resistance, suggesting their involvement in the resistance gene network. This review of our results indicates that TRV-VIGS is an exquisite reverse genetics tool that may provide new insights into the molecular mechanisms underlying plant infection and resistance to infection by begomoviruses.
Figures

Similar articles
-
Plant responses to geminivirus infection: guardians of the plant immunity.Virol J. 2021 Jul 9;18(1):143. doi: 10.1186/s12985-021-01612-1. Virol J. 2021. PMID: 34243802 Free PMC article. Review.
-
Transcriptional reprogramming caused by the geminivirus Tomato yellow leaf curl virus in local or systemic infections in Nicotiana benthamiana.BMC Genomics. 2019 Jul 4;20(1):542. doi: 10.1186/s12864-019-5842-7. BMC Genomics. 2019. PMID: 31272383 Free PMC article.
-
Identification of host genes involved in geminivirus infection using a reverse genetics approach.PLoS One. 2011;6(7):e22383. doi: 10.1371/journal.pone.0022383. Epub 2011 Jul 26. PLoS One. 2011. PMID: 21818318 Free PMC article.
-
Tomato yellow leaf curl virus infection of a resistant tomato line with a silenced sucrose transporter gene LeHT1 results in inhibition of growth, enhanced virus spread, and necrosis.Planta. 2010 Feb;231(3):537-48. doi: 10.1007/s00425-009-1072-6. Epub 2009 Nov 28. Planta. 2010. PMID: 19946703
-
Geminiviruses and Plant Hosts: A Closer Examination of the Molecular Arms Race.Viruses. 2017 Sep 15;9(9):256. doi: 10.3390/v9090256. Viruses. 2017. PMID: 28914771 Free PMC article. Review.
Cited by
-
Insights into the early transcriptomic response against watermelon mosaic virus in melon.BMC Plant Biol. 2024 Jan 20;24(1):58. doi: 10.1186/s12870-024-04745-x. BMC Plant Biol. 2024. PMID: 38245701 Free PMC article.
-
The selection and validation of reference genes for quantitative real-time PCR studies in near-isogenic susceptible and resistant tomato lines, infected with the geminivirus tomato curly stunt virus.PLoS One. 2023 Jul 27;18(7):e0284456. doi: 10.1371/journal.pone.0284456. eCollection 2023. PLoS One. 2023. PMID: 37498814 Free PMC article.
-
Reference Gene Selection for qPCR Analysis in Tomato-Bipartite Begomovirus Interaction and Validation in Additional Tomato-Virus Pathosystems.PLoS One. 2015 Aug 28;10(8):e0136820. doi: 10.1371/journal.pone.0136820. eCollection 2015. PLoS One. 2015. PMID: 26317870 Free PMC article.
-
Quantitative proteomics identifies 38 proteins that are differentially expressed in cucumber in response to cucumber green mottle mosaic virus infection.Virol J. 2015 Dec 15;12:216. doi: 10.1186/s12985-015-0442-x. Virol J. 2015. PMID: 26666291 Free PMC article.
-
Plant responses to geminivirus infection: guardians of the plant immunity.Virol J. 2021 Jul 9;18(1):143. doi: 10.1186/s12985-021-01612-1. Virol J. 2021. PMID: 34243802 Free PMC article. Review.
References
-
- McGregor C.E., Miano D.W., Hoy M., Clark C.A., Rosa G.J.M. Differential gene expression of resistant and susceptible sweetpotato plants after infection with the causal agents of sweet potato virus disease. J. Amer. Soc. Hort. Sci. 2009;134:658–666.
-
- Ascencio-Ibanez J.T., Sozzani R., Lee T.J., Chu T.M., Wolfinger R.D., Cella R., Hanley-Bowdoin L. Global analysis of Arabidopsis gene expression uncovers a complex array of changes impacting pathogen response and cell cycle during geminivirus infection. Plant Physiol. 2008;148:436–454. doi: 10.1104/pp.108.121038. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

