Detection of drug-drug interactions by modeling interaction profile fingerprints
- PMID: 23520498
- PMCID: PMC3592896
- DOI: 10.1371/journal.pone.0058321
Detection of drug-drug interactions by modeling interaction profile fingerprints
Abstract
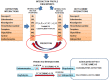
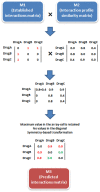
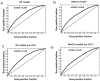
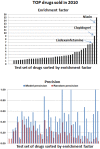
Drug-drug interactions (DDIs) constitute an important problem in postmarketing pharmacovigilance and in the development of new drugs. The effectiveness or toxicity of a medication could be affected by the co-administration of other drugs that share pharmacokinetic or pharmacodynamic pathways. For this reason, a great effort is being made to develop new methodologies to detect and assess DDIs. In this article, we present a novel method based on drug interaction profile fingerprints (IPFs) with successful application to DDI detection. IPFs were generated based on the DrugBank database, which provided 9,454 well-established DDIs as a primary source of interaction data. The model uses IPFs to measure the similarity of pairs of drugs and generates new putative DDIs from the non-intersecting interactions of a pair. We described as part of our analysis the pharmacological and biological effects associated with the putative interactions; for example, the interaction between haloperidol and dicyclomine can cause increased risk of psychosis and tardive dyskinesia. First, we evaluated the method through hold-out validation and then by using four independent test sets that did not overlap with DrugBank. Precision for the test sets ranged from 0.4-0.5 with more than two fold enrichment factor enhancement. In conclusion, we demonstrated the usefulness of the method in pharmacovigilance as a DDI predictor, and created a dataset of potential DDIs, highlighting the etiology or pharmacological effect of the DDI, and providing an exploratory tool to facilitate decision support in DDI detection and patient safety.
Conflict of interest statement
Figures

Similar articles
-
State of the art and development of a drug-drug interaction large scale predictor based on 3D pharmacophoric similarity.Curr Drug Metab. 2014;15(5):490-501. doi: 10.2174/138920021505141126102223. Curr Drug Metab. 2014. PMID: 25431152
-
Drug-drug interaction through molecular structure similarity analysis.J Am Med Inform Assoc. 2012 Nov-Dec;19(6):1066-74. doi: 10.1136/amiajnl-2012-000935. Epub 2012 May 30. J Am Med Inform Assoc. 2012. PMID: 22647690 Free PMC article.
-
Positive-Unlabeled Learning for inferring drug interactions based on heterogeneous attributes.BMC Bioinformatics. 2017 Mar 1;18(1):140. doi: 10.1186/s12859-017-1546-7. BMC Bioinformatics. 2017. PMID: 28249566 Free PMC article.
-
In vitro and in vivo methods to assess pharmacokinetic drug- drug interactions in drug discovery and development.Biopharm Drug Dispos. 2020 Feb;41(1-2):3-31. doi: 10.1002/bdd.2212. Epub 2020 Jan 7. Biopharm Drug Dispos. 2020. PMID: 31778578 Review.
-
Drug-Drug Interactions (DDIs) in Psychiatric Practice, Part 6: Pharmacodynamic Considerations.J Psychiatr Pract. 2019 Jul;25(4):290-297. doi: 10.1097/PRA.0000000000000399. J Psychiatr Pract. 2019. PMID: 31291209 Review.
Cited by
-
Improving Detection of Arrhythmia Drug-Drug Interactions in Pharmacovigilance Data through the Implementation of Similarity-Based Modeling.PLoS One. 2015 Jun 12;10(6):e0129974. doi: 10.1371/journal.pone.0129974. eCollection 2015. PLoS One. 2015. PMID: 26068584 Free PMC article.
-
On the road to explainable AI in drug-drug interactions prediction: A systematic review.Comput Struct Biotechnol J. 2022 Apr 19;20:2112-2123. doi: 10.1016/j.csbj.2022.04.021. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 35832629 Free PMC article. Review.
-
Predicting potential drug-drug interactions by integrating chemical, biological, phenotypic and network data.BMC Bioinformatics. 2017 Jan 5;18(1):18. doi: 10.1186/s12859-016-1415-9. BMC Bioinformatics. 2017. PMID: 28056782 Free PMC article.
-
MSDAFL: molecular substructure-based dual attention feature learning framework for predicting drug-drug interactions.Bioinformatics. 2024 Oct 1;40(10):btae596. doi: 10.1093/bioinformatics/btae596. Bioinformatics. 2024. PMID: 39383521 Free PMC article.
-
A novel efficient drug repurposing framework through drug-disease association data integration using convolutional neural networks.BMC Bioinformatics. 2023 Nov 22;24(1):442. doi: 10.1186/s12859-023-05572-x. BMC Bioinformatics. 2023. PMID: 37993777 Free PMC article.
References
-
- U.S. Food and Drug Administration (FDA). Available: http://www.fda.gov/. Accessed 01 December 2011.
-
- Bjornsson TD, Callaghan JT, Einolf HJ, Fischer V, Gan L, et al. (2003) The conduct of in vitro and in vivo drug-drug interaction studies: A Pharmaceutical Research and Manufacturers of America (PhRMA) perspective. Drug Metabolism and Disposition 31: 815–832. - PubMed
-
- Meinertz T (2001) Mibefradil - a drug which may enhance the propensity for the development of abnormal QT prolongation. Eur Heart J Suppl 3: K89–K92.
-
- Vilar S, Harpaz R, Uriarte E, Santana L, Rabadan R, et al.. (2012) Drug-drug interaction through molecular structure similarity analysis. J Am Med Inform Assoc doi:10.1136/amiajnl-2012-000935. - DOI - PMC - PubMed
-
- Martin YC, Kofron JL, Traphagen LM (2002) Do structurally similar molecules have similar biological activity? J Med Chem 45: 4350–4358. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

