Target-derived neurotrophins coordinate transcription and transport of bclw to prevent axonal degeneration
- PMID: 23516285
- PMCID: PMC3866501
- DOI: 10.1523/JNEUROSCI.3862-12.2013
Target-derived neurotrophins coordinate transcription and transport of bclw to prevent axonal degeneration
Abstract
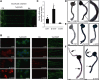
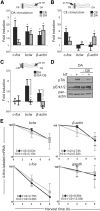
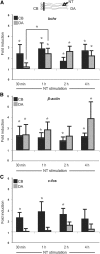
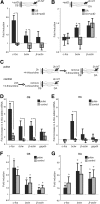
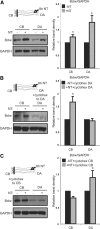
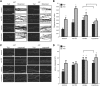
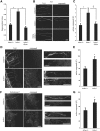
Establishment of neuronal circuitry depends on both formation and refinement of neural connections. During this process, target-derived neurotrophins regulate both transcription and translation to enable selective axon survival or elimination. However, it is not known whether retrograde signaling pathways that control transcription are coordinated with neurotrophin-regulated actions that transpire in the axon. Here we report that target-derived neurotrophins coordinate transcription of the antiapoptotic gene bclw with transport of bclw mRNA to the axon, and thereby prevent axonal degeneration in rat and mouse sensory neurons. We show that neurotrophin stimulation of nerve terminals elicits new bclw transcripts that are immediately transported to the axons and translated into protein. Bclw interacts with Bax and suppresses the caspase6 apoptotic cascade that fosters axonal degeneration. The scope of bclw regulation at the levels of transcription, transport, and translation provides a mechanism whereby sustained neurotrophin stimulation can be integrated over time, so that axonal survival is restricted to neurons connected within a stable circuit.
Figures


Similar articles
-
Paclitaxel Reduces Axonal Bclw to Initiate IP3R1-Dependent Axon Degeneration.Neuron. 2017 Oct 11;96(2):373-386.e6. doi: 10.1016/j.neuron.2017.09.034. Neuron. 2017. PMID: 29024661 Free PMC article.
-
Dynein motors transport activated Trks to promote survival of target-dependent neurons.Nat Neurosci. 2004 Jun;7(6):596-604. doi: 10.1038/nn1242. Epub 2004 May 2. Nat Neurosci. 2004. PMID: 15122257
-
The RNA-binding protein SFPQ orchestrates an RNA regulon to promote axon viability.Nat Neurosci. 2016 May;19(5):690-696. doi: 10.1038/nn.4280. Epub 2016 Mar 28. Nat Neurosci. 2016. PMID: 27019013 Free PMC article.
-
Molecular mechanisms regulating the retrograde axonal transport of neurotrophins.Brain Res Brain Res Rev. 2000 Sep;33(2-3):169-78. doi: 10.1016/s0165-0173(00)00028-x. Brain Res Brain Res Rev. 2000. PMID: 11011064 Review.
-
Retrograde axonal transport of neurotrophins: differences between neuronal populations and implications for motor neuron disease.Immunol Cell Biol. 1998 Oct;76(5):419-23. doi: 10.1046/j.1440-1711.1998.00767.x. Immunol Cell Biol. 1998. PMID: 9797461 Review.
Cited by
-
DLK-dependent axonal mitochondrial fission drives degeneration following axotomy.bioRxiv [Preprint]. 2024 Jun 4:2023.01.30.526132. doi: 10.1101/2023.01.30.526132. bioRxiv. 2024. Update in: Nat Commun. 2024 Dec 30;15(1):10806. doi: 10.1038/s41467-024-54982-9. PMID: 36778383 Free PMC article. Updated. Preprint.
-
Physiological functions of non-apoptotic caspase activity in the nervous system.Semin Cell Dev Biol. 2018 Oct;82:127-136. doi: 10.1016/j.semcdb.2017.11.037. Epub 2017 Dec 7. Semin Cell Dev Biol. 2018. PMID: 29199140 Free PMC article. Review.
-
Axonal mRNA localization and translation: local events with broad roles.Cell Mol Life Sci. 2021 Dec;78(23):7379-7395. doi: 10.1007/s00018-021-03995-4. Epub 2021 Oct 26. Cell Mol Life Sci. 2021. PMID: 34698881 Free PMC article. Review.
-
Looking for answers far away from the soma-the (un)known axonal functions of TDP-43, and their contribution to early NMJ disruption in ALS.Mol Neurodegener. 2023 May 31;18(1):35. doi: 10.1186/s13024-023-00623-6. Mol Neurodegener. 2023. PMID: 37259156 Free PMC article. Review.
-
Binding and transport of SFPQ-RNA granules by KIF5A/KLC1 motors promotes axon survival.J Cell Biol. 2021 Jan 4;220(1):e202005051. doi: 10.1083/jcb.202005051. J Cell Biol. 2021. PMID: 33284322 Free PMC article.
References
-
- Andreassi C, Riccio A. To localize or not to localize: mRNA fate is in 3′UTR ends. Trends Cell Biol. 2009;19:465–474. - PubMed
-
- Andreassi C, Zimmermann C, Mitter R, Fusco S, Devita S, Saiardi A, Riccio A. An NGF-responsive element targets myo-inositol monophosphatase-1 mRNA to sympathetic neuron axons. Nat Neurosci. 2010;13:291–301. - PubMed
-
- Brumovsky P, Villar MJ, Hökfelt T. Tyrosine hydroxylase is expressed in a subpopulation of small dorsal root ganglion neurons in the adult mouse. Exp Neurol. 2006;200:153–165. - PubMed
-
- Campenot RB. Development of sympathetic neurons in compartmentalized cultures: II. Local control of neurite survival by nerve growth factor. Dev Biol. 1982;93:13–21. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
