Sumoylation of Krüppel-like factor 4 inhibits pluripotency induction but promotes adipocyte differentiation
- PMID: 23515309
- PMCID: PMC3642324
- DOI: 10.1074/jbc.M113.465443
Sumoylation of Krüppel-like factor 4 inhibits pluripotency induction but promotes adipocyte differentiation
Abstract
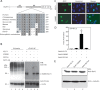
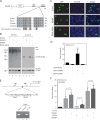
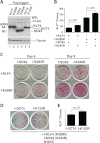
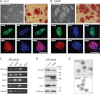
Ectopic expression of transcription factors has been shown to reprogram somatic cells into induced pluripotent stem (iPS) cells. It remains largely unexplored how this process is regulated by post-translational modifications. Several reprogramming factors possess conserved sumoylation sites, so we investigated whether and how this modification regulates reprogramming of fibroblasts into iPS cells. Substitution of the sole sumoylation site of the Krüppel-like factor (KLF4), a well known reprogramming factor, promoted iPS cell formation. In comparison, much smaller effects on reprogramming were observed for sumoylation-deficient mutants of SOX2 and OCT4, two other classical reprogramming factors. We also analyzed KLF2, a KLF4 homolog and a member of the KLF family of transcription factors with a known role in reprogramming. KLF2 was sumoylated at two conserved neighboring motifs, but substitution of the key lysine residues only stimulated reprogramming slightly. KLF5 is another KLF member with an established link to embryonic stem cell pluripotency. Interestingly, although it was much more efficiently sumoylated than either KLF2 or KLF4, KLF5 was inactive in reprogramming, and its sumoylation was not responsible for this deficiency. Furthermore, sumoylation of KLF4 but not KLF2 or KLF5 stimulated adipocyte differentiation. These results thus demonstrate the importance KLF4 sumoylation in regulating pluripotency and adipocyte differentiation.
Keywords: Adipocyte; Cell Differentiation; KLF2; KLF4; KLF5; Reprogramming; Sumoylation; Transcription Factors; iPS Cells.
Figures
Similar articles
-
Klf4 interacts directly with Oct4 and Sox2 to promote reprogramming.Stem Cells. 2009 Dec;27(12):2969-78. doi: 10.1002/stem.231. Stem Cells. 2009. PMID: 19816951
-
MicroRNA-302 increases reprogramming efficiency via repression of NR2F2.Stem Cells. 2013 Feb;31(2):259-68. doi: 10.1002/stem.1278. Stem Cells. 2013. PMID: 23136034 Free PMC article.
-
Critical POU domain residues confer Oct4 uniqueness in somatic cell reprogramming.Sci Rep. 2016 Feb 15;6:20818. doi: 10.1038/srep20818. Sci Rep. 2016. PMID: 26877091 Free PMC article.
-
Biological importance of OCT transcription factors in reprogramming and development.Exp Mol Med. 2021 Jun;53(6):1018-1028. doi: 10.1038/s12276-021-00637-4. Epub 2021 Jun 11. Exp Mol Med. 2021. PMID: 34117345 Free PMC article. Review.
-
Choices for Induction of Pluripotency: Recent Developments in Human Induced Pluripotent Stem Cell Reprogramming Strategies.Stem Cell Rev Rep. 2016 Feb;12(1):54-72. doi: 10.1007/s12015-015-9622-8. Stem Cell Rev Rep. 2016. PMID: 26424535 Free PMC article. Review.
Cited by
-
F-box protein FBXO22 mediates polyubiquitination and degradation of KLF4 to promote hepatocellular carcinoma progression.Oncotarget. 2015 Sep 8;6(26):22767-75. doi: 10.18632/oncotarget.4082. Oncotarget. 2015. PMID: 26087183 Free PMC article.
-
The Roles of SUMO in Metabolic Regulation.Adv Exp Med Biol. 2017;963:143-168. doi: 10.1007/978-3-319-50044-7_9. Adv Exp Med Biol. 2017. PMID: 28197911 Free PMC article. Review.
-
Not So Slim Anymore-Evidence for the Role of SUMO in the Regulation of Lipid Metabolism.Biomolecules. 2020 Aug 6;10(8):1154. doi: 10.3390/biom10081154. Biomolecules. 2020. PMID: 32781719 Free PMC article. Review.
-
Mice lacking α-tubulin acetyltransferase 1 are viable but display α-tubulin acetylation deficiency and dentate gyrus distortion.J Biol Chem. 2013 Jul 12;288(28):20334-50. doi: 10.1074/jbc.M113.464792. Epub 2013 May 28. J Biol Chem. 2013. PMID: 23720746 Free PMC article.
-
Effect of Temperature and Selection for Growth on Intracellular Lipid Accumulation and Adipogenic Gene Expression in Turkey Pectoralis Major Muscle Satellite Cells.Front Physiol. 2021 Jun 1;12:667814. doi: 10.3389/fphys.2021.667814. eCollection 2021. Front Physiol. 2021. PMID: 34140894 Free PMC article.
References
-
- Gill G. (2004) SUMO and ubiquitin in the nucleus: different functions, similar mechanisms? Genes Dev. 18, 2046–2059 - PubMed
-
- Geiss-Friedlander R., Melchior F. (2007) Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell Biol. 8, 947–956 - PubMed
-
- Nacerddine K., Lehembre F., Bhaumik M., Artus J., Cohen-Tannoudji M., Babinet C., Pandolfi P. P., Dejean A. (2005) The SUMO pathway is essential for nuclear integrity and chromosome segregation in mice. Dev. Cell 9, 769–779 - PubMed
-
- Hay R. T. (2005) SUMO: a history of modification. Mol. Cell 18, 1–12 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

