Downregulation of Nipah virus N mRNA occurs through interaction between its 3' untranslated region and hnRNP D
- PMID: 23514888
- PMCID: PMC3676090
- DOI: 10.1128/JVI.02495-12
Downregulation of Nipah virus N mRNA occurs through interaction between its 3' untranslated region and hnRNP D
Abstract
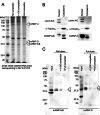
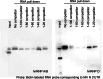
Nipah virus (NiV) is a nonsegmented, single-stranded, negative-sense RNA virus belonging to the genus Henipavirus, family Paramyxoviridae. NiV causes acute encephalitis and respiratory disease in humans, is associated with high mortality, and poses a threat in southern Asia. The genomes of henipaviruses are about 18,246 nucleotides (nt) long, which is longer than those of other paramyxoviruses (around 15,384 nt). This difference is caused by the noncoding RNA region, particularly the 3' untranslated region (UTR), which occupies more than half of the noncoding RNA region. To determine the function(s) of the NiV noncoding RNA region, we investigated the effects of NiV 3' UTRs on reporter gene expression. The NiV N 3' UTR (nt 1 to 100) demonstrated strong repressor activity associated with hnRNP D protein binding to that region. Mutation of the hnRNP D binding site or knockdown of hnRNP D resulted in increased expression of the NiV N 3' UTR reporter. Our findings suggest that NiV N expression is repressed by hnRNP D through the NiV N 3' UTR and demonstrate the involvement of posttranscriptional regulation in the NiV life cycle. To the best of our knowledge, this provides the first report of the functions of the NiV noncoding RNA region.
Figures


Similar articles
-
The critical role of mRNA destabilizing protein heterogeneous nuclear ribonucleoprotein d in 3' untranslated region-mediated decay of low-density lipoprotein receptor mRNA in liver tissue.Arterioscler Thromb Vasc Biol. 2014 Jan;34(1):8-16. doi: 10.1161/ATVBAHA.112.301131. Epub 2013 Oct 24. Arterioscler Thromb Vasc Biol. 2014. PMID: 24158514 Free PMC article.
-
Interplay between hnRNP A1 and a cis-acting element in the 3' UTR of CYP2A5 mRNA is central for high expression of the gene.FEBS Lett. 2003 Jan 30;535(1-3):147-52. doi: 10.1016/s0014-5793(02)03893-0. FEBS Lett. 2003. PMID: 12560094
-
Eukaryotic elongation factor 1-beta interacts with the 5' untranslated region of the M gene of Nipah virus to promote mRNA translation.Arch Virol. 2016 Sep;161(9):2361-8. doi: 10.1007/s00705-016-2903-y. Epub 2016 May 28. Arch Virol. 2016. PMID: 27236461
-
Third Helical Domain of the Nipah Virus Fusion Glycoprotein Modulates both Early and Late Steps in the Membrane Fusion Cascade.J Virol. 2020 Sep 15;94(19):e00644-20. doi: 10.1128/JVI.00644-20. Print 2020 Sep 15. J Virol. 2020. PMID: 32669342 Free PMC article.
-
Possible high risk of transmission of the Nipah virus in South and South East Asia: a review.Trop Med Health. 2023 Aug 10;51(1):44. doi: 10.1186/s41182-023-00535-7. Trop Med Health. 2023. PMID: 37559114 Free PMC article. Review.
Cited by
-
mRNA decay factor AUF1 binds the internal ribosomal entry site of enterovirus 71 and inhibits virus replication.PLoS One. 2014 Jul 31;9(7):e103827. doi: 10.1371/journal.pone.0103827. eCollection 2014. PLoS One. 2014. PMID: 25077793 Free PMC article.
-
Methylenetetrahydrofolate reductase polymorphisms at 3'-untranslated region are associated with susceptibility to preterm birth.Transl Pediatr. 2015 Jan;4(1):57-62. doi: 10.3978/j.issn.2224-4336.2015.01.02. Transl Pediatr. 2015. PMID: 26835361 Free PMC article.
-
Genome-wide transposon mutagenesis of paramyxoviruses reveals constraints on genomic plasticity.PLoS Pathog. 2020 Oct 9;16(10):e1008877. doi: 10.1371/journal.ppat.1008877. eCollection 2020 Oct. PLoS Pathog. 2020. PMID: 33035269 Free PMC article.
-
Arginine methylation enhances the RNA chaperone activity of the West Nile virus host factor AUF1 p45.RNA. 2016 Oct;22(10):1574-91. doi: 10.1261/rna.055269.115. Epub 2016 Aug 12. RNA. 2016. PMID: 27520967 Free PMC article.
-
Protracted molecular dynamics and secondary structure introspection to identify dual-target inhibitors of Nipah virus exerting approved small molecules repurposing.Sci Rep. 2024 Feb 14;14(1):3696. doi: 10.1038/s41598-024-54281-9. Sci Rep. 2024. PMID: 38355980 Free PMC article.
References
-
- Anonymous 2004. Nipah encephalitis outbreak over wide area of western Bangladesh, 2004. Health Sci. Bull. 2:7–11
-
- Chua KB, Bellini WJ, Rota PA, Harcourt BH, Tamin A, Lam SK, Ksiazek TG, Rollin PE, Zaki SR, Shieh W, Goldsmith CS, Gubler DJ, Roehrig JT, Eaton B, Gould AR, Olson J, Field H, Daniels P, Ling AE, Peters CJ, Anderson LJ, Mahy BW. 2000. Nipah virus: a recently emergent deadly paramyxovirus. Science 288:1432–1435 - PubMed
-
- Harcourt BH, Tamin A, Ksiazek TG, Rollin PE, Anderson LJ, Bellini WJ, Rota PA. 2000. Molecular characterization of Nipah virus, a newly emergent paramyxovirus. Virology 271:334–349 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

