Assembly of the 20S proteasome
- PMID: 23507199
- PMCID: PMC3752329
- DOI: 10.1016/j.bbamcr.2013.03.008
Assembly of the 20S proteasome
Abstract
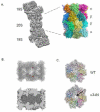
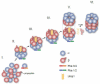
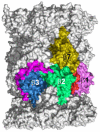
The proteasome is a cellular protease responsible for the selective degradation of the majority of the intracellular proteome. It recognizes, unfolds, and cleaves proteins that are destined for removal, usually by prior attachment to polymers of ubiquitin. This macromolecular machine is composed of two subcomplexes, the 19S regulatory particle (RP) and the 20S core particle (CP), which together contain at least 33 different and precisely positioned subunits. How these subunits assemble into functional complexes is an area of active exploration. Here we describe the current status of studies on the assembly of the 20S proteasome (CP). The 28-subunit CP is found in all three domains of life and its cylindrical stack of four heptameric rings is well conserved. Though several CP subunits possess self-assembly properties, a consistent theme in recent years has been the need for dedicated assembly chaperones that promote on-pathway assembly. To date, a minimum of three accessory factors have been implicated in aiding the construction of the 20S proteasome. These chaperones interact with different assembling proteasomal precursors and usher subunits into specific slots in the growing structure. This review will focus largely on chaperone-dependent CP assembly and its regulation. This article is part of a Special Issue entitled: Ubiquitin-Proteasome System. Guest Editors: Thomas Sommer and Dieter H. Wolf.
Keywords: Assembly factors; Macromolecular complex assembly; Proteasome; Proteolysis; Ubiquitin-proteasome system.
Copyright © 2013 Elsevier B.V. All rights reserved.
Figures

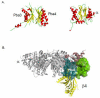
Similar articles
-
Chaperone-assisted assembly of the proteasome core particle.Biochem Soc Trans. 2010 Feb;38(Pt 1):29-33. doi: 10.1042/BST0380029. Biochem Soc Trans. 2010. PMID: 20074030
-
Order of the proteasomal ATPases and eukaryotic proteasome assembly.Cell Biochem Biophys. 2011 Jun;60(1-2):13-20. doi: 10.1007/s12013-011-9178-4. Cell Biochem Biophys. 2011. PMID: 21461838 Free PMC article. Review.
-
PACemakers of proteasome core particle assembly.Structure. 2008 Sep 10;16(9):1296-304. doi: 10.1016/j.str.2008.07.001. Structure. 2008. PMID: 18786393 Review.
-
Chaperone-driven proteasome assembly.Biochem Soc Trans. 2008 Oct;36(Pt 5):807-12. doi: 10.1042/BST0360807. Biochem Soc Trans. 2008. PMID: 18793141 Review.
-
A heterodimeric complex that promotes the assembly of mammalian 20S proteasomes.Nature. 2005 Oct 27;437(7063):1381-5. doi: 10.1038/nature04106. Nature. 2005. PMID: 16251969
Cited by
-
The role of the ubiquitin proteasome system in cerebellar development and medulloblastoma.Mol Brain. 2015 Oct 17;8(1):64. doi: 10.1186/s13041-015-0155-5. Mol Brain. 2015. PMID: 26475605 Free PMC article. Review.
-
Gates, Channels, and Switches: Elements of the Proteasome Machine.Trends Biochem Sci. 2016 Jan;41(1):77-93. doi: 10.1016/j.tibs.2015.10.009. Epub 2015 Nov 28. Trends Biochem Sci. 2016. PMID: 26643069 Free PMC article. Review.
-
Drug Combination Studies of Isoquinolinone AM12 with Curcumin or Quercetin: A New Combination Strategy to Synergistically Inhibit 20S Proteasome.Int J Mol Sci. 2024 Oct 4;25(19):10708. doi: 10.3390/ijms251910708. Int J Mol Sci. 2024. PMID: 39409037 Free PMC article.
-
Maturation of the proteasome core particle induces an affinity switch that controls regulatory particle association.Nat Commun. 2015 Mar 16;6:6384. doi: 10.1038/ncomms7384. Nat Commun. 2015. PMID: 25812915 Free PMC article.
-
Bacterial Proteasomes.Annu Rev Microbiol. 2015;69:109-27. doi: 10.1146/annurev-micro-091014-104201. Annu Rev Microbiol. 2015. PMID: 26488274 Free PMC article. Review.
References
-
- Hochstrasser M. Ubiquitin-dependent protein degradation. Annu. Rev. Genet. 1996;30:405–439. - PubMed
-
- Ciechanover A, Schwartz AL. The ubiquitin system: pathogenesis of human diseases and drug targeting. Biochim. Biophys. Acta. 2004;1695:3–17. - PubMed
-
- Ciechanover A. Proteolysis: from the lysosome to ubiquitin and the proteasome. Nat. Rev. Mol. Cell Biol. 2005;6:79–87. - PubMed
-
- Komander D, Rape M. The ubiquitin code. Annu. Rev. Biochem. 2012;81:203–229. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

