Crystal structure-based exploration of the important role of Arg106 in the RNA-binding domain of human coronavirus OC43 nucleocapsid protein
- PMID: 23501675
- PMCID: PMC3774783
- DOI: 10.1016/j.bbapap.2013.03.003
Crystal structure-based exploration of the important role of Arg106 in the RNA-binding domain of human coronavirus OC43 nucleocapsid protein
Abstract
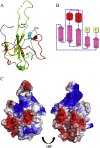
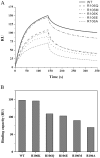
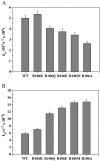
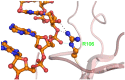
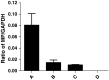
Human coronavirus OC43 (HCoV-OC43) is a causative agent of the common cold. The nucleocapsid (N) protein, which is a major structural protein of CoVs, binds to the viral RNA genome to form the virion core and results in the formation of the ribonucleoprotein (RNP) complex. We have solved the crystal structure of the N-terminal domain of HCoV-OC43 N protein (N-NTD) (residues 58 to 195) to a resolution of 2.0Å. The HCoV-OC43 N-NTD is a single domain protein composed of a five-stranded β-sheet core and a long extended loop, similar to that observed in the structures of N-NTDs from other coronaviruses. The positively charged loop of the HCoV-OC43 N-NTD contains a structurally well-conserved positively charged residue, R106. To assess the role of R106 in RNA binding, we undertook a series of site-directed mutagenesis experiments and docking simulations to characterize the interaction between R106 and RNA. The results show that R106 plays an important role in the interaction between the N protein and RNA. In addition, we showed that, in cells transfected with plasmids that encoded the mutant (R106A) N protein and infected with virus, the level of the matrix protein gene was decreased by 7-fold compared to cells that were transfected with the wild-type N protein. This finding suggests that R106, by enhancing binding of the N protein to viral RNA plays a critical role in the viral replication. The results also indicate that the strength of N protein/RNA interactions is critical for HCoV-OC43 replication.
Crown Copyright © 2013. Published by Elsevier B.V. All rights reserved.
Figures


Similar articles
-
Elucidation of the stability and functional regions of the human coronavirus OC43 nucleocapsid protein.Protein Sci. 2009 Nov;18(11):2209-18. doi: 10.1002/pro.225. Protein Sci. 2009. PMID: 19691129 Free PMC article.
-
Crystallization and preliminary X-ray diffraction analysis of the N-terminal domain of human coronavirus OC43 nucleocapsid protein.Acta Crystallogr Sect F Struct Biol Cryst Commun. 2010 Jul 1;66(Pt 7):815-8. doi: 10.1107/S1744309110017616. Epub 2010 Jun 24. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2010. PMID: 20606281 Free PMC article.
-
Coronavirus N protein N-terminal domain (NTD) specifically binds the transcriptional regulatory sequence (TRS) and melts TRS-cTRS RNA duplexes.J Mol Biol. 2009 Dec 4;394(3):544-57. doi: 10.1016/j.jmb.2009.09.040. Epub 2009 Sep 24. J Mol Biol. 2009. PMID: 19782089 Free PMC article.
-
Properties of Coronavirus and SARS-CoV-2.Malays J Pathol. 2020 Apr;42(1):3-11. Malays J Pathol. 2020. PMID: 32342926 Review.
-
The SARS coronavirus nucleocapsid protein--forms and functions.Antiviral Res. 2014 Mar;103:39-50. doi: 10.1016/j.antiviral.2013.12.009. Epub 2014 Jan 11. Antiviral Res. 2014. PMID: 24418573 Free PMC article. Review.
Cited by
-
Targeting protein-protein interaction interfaces with antiviral N protein inhibitor in SARS-CoV-2.Biophys J. 2024 Feb 20;123(4):478-488. doi: 10.1016/j.bpj.2024.01.013. Epub 2024 Jan 17. Biophys J. 2024. PMID: 38234090
-
Cell surface nucleocapsid protein expression: A betacoronavirus immunomodulatory strategy.Proc Natl Acad Sci U S A. 2023 Jul 11;120(28):e2304087120. doi: 10.1073/pnas.2304087120. Epub 2023 Jul 3. Proc Natl Acad Sci U S A. 2023. PMID: 37399385 Free PMC article.
-
Cell Surface Nucleocapsid Protein Expression: A Betacoronavirus Immunomodulatory Strategy.bioRxiv [Preprint]. 2023 Feb 27:2023.02.24.529952. doi: 10.1101/2023.02.24.529952. bioRxiv. 2023. Update in: Proc Natl Acad Sci U S A. 2023 Jul 11;120(28):e2304087120. doi: 10.1073/pnas.2304087120 PMID: 36993159 Free PMC article. Updated. Preprint.
-
Structural basis of RNA recognition by the SARS-CoV-2 nucleocapsid phosphoprotein.PLoS Pathog. 2020 Dec 2;16(12):e1009100. doi: 10.1371/journal.ppat.1009100. eCollection 2020 Dec. PLoS Pathog. 2020. PMID: 33264373 Free PMC article.
-
In silico screening of hundred phytocompounds of ten medicinal plants as potential inhibitors of nucleocapsid phosphoprotein of COVID-19: an approach to prevent virus assembly.J Biomol Struct Dyn. 2021 Nov;39(18):7017-7034. doi: 10.1080/07391102.2020.1804457. Epub 2020 Aug 27. J Biomol Struct Dyn. 2021. PMID: 32851912 Free PMC article.
References
-
- Wenzel R.P., Hendley J.O., Davies J.A., Gwaltney J.M., Jr. Coronavirus infections in military recruits. Three-year study with coronavirus strains OC43 and 229E. Am. Rev. Respir. Dis. 1974;109:621–624. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

