Archaeal proteasomes and sampylation
- PMID: 23479445
- PMCID: PMC3936409
- DOI: 10.1007/978-94-007-5940-4_11
Archaeal proteasomes and sampylation
Abstract
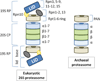
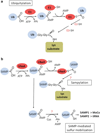
Archaea contain, both a functional proteasome and an ubiquitin-like protein conjugation system (termed sampylation) that is related to the ubiquitin proteasome system (UPS) of eukaryotes. Archaeal proteasomes have served as excellent models for understanding how proteins are degraded by the central energy-dependent proteolytic machine of eukaryotes, the 26S proteasome. While sampylation has only recently been discovered, it is thought to be linked to proteasome-mediated degradation in archaea. Unlike eukaryotes, sampylation only requires an E1 enzyme homolog of the E1-E2-E3 ubiquitylation cascade to mediate protein conjugation. Furthermore, recent evidence suggests that archaeal and eurkaryotic E1 enzyme homologs can serve dual roles in mediating protein conjugation and activating sulfur for incorporation into biomolecules. The focus of this book chapter is the energy-dependent proteasome and sampylation systems of Archaea.
Figures


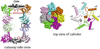


Similar articles
-
Ubiquitin-Like Proteasome System Represents a Eukaryotic-Like Pathway for Targeted Proteolysis in Archaea.mBio. 2016 May 17;7(3):e00379-16. doi: 10.1128/mBio.00379-16. mBio. 2016. PMID: 27190215 Free PMC article.
-
Prokaryotic proteasomes: nanocompartments of degradation.J Mol Microbiol Biotechnol. 2013;23(4-5):321-34. doi: 10.1159/000351348. Epub 2013 Aug 5. J Mol Microbiol Biotechnol. 2013. PMID: 23920495 Free PMC article. Review.
-
Rhodanese-Like Domain Protein UbaC and Its Role in Ubiquitin-Like Protein Modification and Sulfur Mobilization in Archaea.J Bacteriol. 2019 Jul 10;201(15):e00254-19. doi: 10.1128/JB.00254-19. Print 2019 Aug 1. J Bacteriol. 2019. PMID: 31085691 Free PMC article.
-
Prokaryotic ubiquitin-like protein modification.Annu Rev Microbiol. 2014;68:155-75. doi: 10.1146/annurev-micro-091313-103447. Epub 2014 May 29. Annu Rev Microbiol. 2014. PMID: 24995873 Free PMC article. Review.
-
Ubiquitin-like proteins and their roles in archaea.Trends Microbiol. 2013 Jan;21(1):31-8. doi: 10.1016/j.tim.2012.09.006. Epub 2012 Nov 8. Trends Microbiol. 2013. PMID: 23140889 Free PMC article. Review.
Cited by
-
The pup-proteasome system of Mycobacterium tuberculosis.Subcell Biochem. 2013;66:267-95. doi: 10.1007/978-94-007-5940-4_10. Subcell Biochem. 2013. PMID: 23479444 Free PMC article. Review.
-
On the origin of the nucleus: a hypothesis.Microbiol Mol Biol Rev. 2023 Dec 20;87(4):e0018621. doi: 10.1128/mmbr.00186-21. Epub 2023 Nov 29. Microbiol Mol Biol Rev. 2023. PMID: 38018971 Free PMC article. Review.
-
Backbone NMR resonance assignment of the intrinsically disordered UBact protein from Nitrospira nitrosa.Biomol NMR Assign. 2022 Apr;16(1):129-134. doi: 10.1007/s12104-022-10070-x. Epub 2022 Feb 2. Biomol NMR Assign. 2022. PMID: 35107780 Free PMC article.
-
Multidomain ribosomal protein trees and the planctobacterial origin of neomura (eukaryotes, archaebacteria).Protoplasma. 2020 May;257(3):621-753. doi: 10.1007/s00709-019-01442-7. Epub 2020 Jan 3. Protoplasma. 2020. PMID: 31900730 Free PMC article. Review.
-
Molecular Factors of Hypochlorite Tolerance in the Hypersaline Archaeon Haloferax volcanii.Genes (Basel). 2018 Nov 20;9(11):562. doi: 10.3390/genes9110562. Genes (Basel). 2018. PMID: 30463375 Free PMC article.
References
-
- Gottesman S. Regulation by proteolysis: developmental switches. Curr Opin Microbiol. 1999;2(2):142–147. - PubMed
-
- Wolf DH, Hilt W. The proteasome: a proteolytic nanomachine of cell regulation and waste disposal. Biochim Biophys Acta. 2004;1695(1–3):19–31. - PubMed
-
- Lupas A, Flanagan JM, Tamura T, Baumeister W. Self-compartmentalizing proteases. Trends Biochem Sci. 1997;22(10):399–404. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

