STING recognition of cytoplasmic DNA instigates cellular defense
- PMID: 23478444
- PMCID: PMC3881179
- DOI: 10.1016/j.molcel.2013.01.039
STING recognition of cytoplasmic DNA instigates cellular defense
Abstract
How the cell recognizes cytosolic DNA including DNA-based microbes to trigger host-defense-related gene activation remains to be fully resolved. Here, we demonstrate that STING (stimulator of interferon genes), an endoplasmic reticulum translocon-associated transmembrane protein, acts to detect cytoplasmic DNA species. STING homodimers were able to complex with self- (apoptotic, necrotic) or pathogen-related ssDNA and dsDNA and were indispensible for HSV-1-mediated transcriptional activation of a wide array of innate immune and proinflammatory genes in addition to type I IFN. Our data indicate that STING instigates cytoplasmic DNA-mediated cellular defense gene transcription and facilitates adoptive responses that are required for protection of the host. In contrast, chronic STING activation may manifest inflammatory responses and possibly autoimmune disease triggered by self-DNA.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures

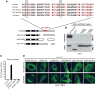
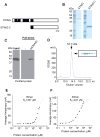
Comment in
-
How do cells sense foreign DNA? A new outlook on the function of STING.Mol Cell. 2013 Apr 11;50(1):1-2. doi: 10.1016/j.molcel.2013.03.024. Mol Cell. 2013. PMID: 23582257
Similar articles
-
STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity.Nature. 2009 Oct 8;461(7265):788-92. doi: 10.1038/nature08476. Epub 2009 Sep 23. Nature. 2009. PMID: 19776740 Free PMC article.
-
The GRA15 protein from Toxoplasma gondii enhances host defense responses by activating the interferon stimulator STING.J Biol Chem. 2019 Nov 8;294(45):16494-16508. doi: 10.1074/jbc.RA119.009172. Epub 2019 Aug 15. J Biol Chem. 2019. PMID: 31416833 Free PMC article.
-
Herpesvirus Genome Recognition Induced Acetylation of Nuclear IFI16 Is Essential for Its Cytoplasmic Translocation, Inflammasome and IFN-β Responses.PLoS Pathog. 2015 Jul 2;11(7):e1005019. doi: 10.1371/journal.ppat.1005019. eCollection 2015 Jul. PLoS Pathog. 2015. PMID: 26134128 Free PMC article.
-
Cytoplasmic DNA innate immune pathways.Immunol Rev. 2011 Sep;243(1):99-108. doi: 10.1111/j.1600-065X.2011.01051.x. Immunol Rev. 2011. PMID: 21884170 Review.
-
Innate Immune Response to Cytoplasmic DNA: Mechanisms and Diseases.Annu Rev Immunol. 2020 Apr 26;38:79-98. doi: 10.1146/annurev-immunol-070119-115052. Epub 2019 Dec 4. Annu Rev Immunol. 2020. PMID: 31800327 Review.
Cited by
-
Cell Free DNA of Tumor Origin Induces a 'Metastatic' Expression Profile in HT-29 Cancer Cell Line.PLoS One. 2015 Jul 2;10(7):e0131699. doi: 10.1371/journal.pone.0131699. eCollection 2015. PLoS One. 2015. PMID: 26133168 Free PMC article.
-
Chemical and Biomolecular Strategies for STING Pathway Activation in Cancer Immunotherapy.Chem Rev. 2022 Mar 23;122(6):5977-6039. doi: 10.1021/acs.chemrev.1c00750. Epub 2022 Feb 2. Chem Rev. 2022. PMID: 35107989 Free PMC article. Review.
-
STING pathway agonism as a cancer therapeutic.Immunol Rev. 2019 Jul;290(1):24-38. doi: 10.1111/imr.12765. Immunol Rev. 2019. PMID: 31355488 Free PMC article. Review.
-
STING activation in cancer immunotherapy.Theranostics. 2019 Oct 15;9(25):7759-7771. doi: 10.7150/thno.37574. eCollection 2019. Theranostics. 2019. PMID: 31695799 Free PMC article. Review.
-
Ovarian Cancer Cells Commonly Exhibit Defective STING Signaling Which Affects Sensitivity to Viral Oncolysis.Mol Cancer Res. 2019 Apr;17(4):974-986. doi: 10.1158/1541-7786.MCR-18-0504. Epub 2018 Dec 26. Mol Cancer Res. 2019. PMID: 30587523 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

