Super-resolution microscopy reveals specific recruitment of HIV-1 envelope proteins to viral assembly sites dependent on the envelope C-terminal tail
- PMID: 23468635
- PMCID: PMC3585150
- DOI: 10.1371/journal.ppat.1003198
Super-resolution microscopy reveals specific recruitment of HIV-1 envelope proteins to viral assembly sites dependent on the envelope C-terminal tail
Abstract
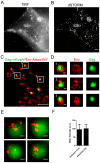
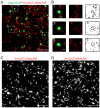
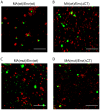
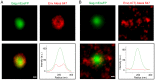
The inner structural Gag proteins and the envelope (Env) glycoproteins of human immunodeficiency virus (HIV-1) traffic independently to the plasma membrane, where they assemble the nascent virion. HIV-1 carries a relatively low number of glycoproteins in its membrane, and the mechanism of Env recruitment and virus incorporation is incompletely understood. We employed dual-color super-resolution microscopy visualizing Gag assembly sites and HIV-1 Env proteins in virus-producing and in Env expressing cells. Distinctive HIV-1 Gag assembly sites were readily detected and were associated with Env clusters that always extended beyond the actual Gag assembly site and often showed enrichment at the periphery and surrounding the assembly site. Formation of these Env clusters depended on the presence of other HIV-1 proteins and on the long cytoplasmic tail (CT) of Env. CT deletion, a matrix mutation affecting Env incorporation or Env expression in the absence of other HIV-1 proteins led to much smaller Env clusters, which were not enriched at viral assembly sites. These results show that Env is recruited to HIV-1 assembly sites in a CT-dependent manner, while Env(ΔCT) appears to be randomly incorporated. The observed Env accumulation surrounding Gag assemblies, with a lower density on the actual bud, could facilitate viral spread in vivo. Keeping Env molecules on the nascent virus low may be important for escape from the humoral immune response, while cell-cell contacts mediated by surrounding Env molecules could promote HIV-1 transmission through the virological synapse.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
Probing Gag-Env dynamics at HIV-1 assembly sites using live-cell microscopy.J Virol. 2024 Sep 17;98(9):e0064924. doi: 10.1128/jvi.00649-24. Epub 2024 Aug 13. J Virol. 2024. PMID: 39136462 Free PMC article.
-
HIV-1 Gag, Envelope, and Extracellular Determinants Cooperate To Regulate the Stability and Turnover of Virological Synapses.J Virol. 2016 Jun 24;90(14):6583-6597. doi: 10.1128/JVI.00600-16. Print 2016 Jul 15. J Virol. 2016. PMID: 27170746 Free PMC article.
-
Clustering and mobility of HIV-1 Env at viral assembly sites predict its propensity to induce cell-cell fusion.J Virol. 2013 Jul;87(13):7516-25. doi: 10.1128/JVI.00790-13. Epub 2013 May 1. J Virol. 2013. PMID: 23637402 Free PMC article.
-
The Interplay between HIV-1 Gag Binding to the Plasma Membrane and Env Incorporation.Viruses. 2020 May 16;12(5):548. doi: 10.3390/v12050548. Viruses. 2020. PMID: 32429351 Free PMC article. Review.
-
The role of matrix in HIV-1 envelope glycoprotein incorporation.Trends Microbiol. 2014 Jul;22(7):372-8. doi: 10.1016/j.tim.2014.04.012. Epub 2014 Jun 2. Trends Microbiol. 2014. PMID: 24933691 Free PMC article. Review.
Cited by
-
Blockage of CD59 Function Restores Activities of Neutralizing and Nonneutralizing Antibodies in Triggering Antibody-Dependent Complement-Mediated Lysis of HIV-1 Virions and Provirus-Activated Latently Infected Cells.J Virol. 2015 Sep;89(18):9393-406. doi: 10.1128/JVI.01614-15. Epub 2015 Jul 1. J Virol. 2015. PMID: 26136568 Free PMC article.
-
The cytoplasmic tail of retroviral envelope glycoproteins.Prog Mol Biol Transl Sci. 2015;129:253-84. doi: 10.1016/bs.pmbts.2014.10.009. Epub 2014 Dec 1. Prog Mol Biol Transl Sci. 2015. PMID: 25595807 Free PMC article. Review.
-
Single-Virus Tracking: From Imaging Methodologies to Virological Applications.Chem Rev. 2020 Feb 12;120(3):1936-1979. doi: 10.1021/acs.chemrev.9b00692. Epub 2020 Jan 17. Chem Rev. 2020. PMID: 31951121 Free PMC article. Review.
-
Second site reversion of HIV-1 envelope protein baseplate mutations maps to the matrix protein.J Virol. 2024 Feb 20;98(2):e0174223. doi: 10.1128/jvi.01742-23. Epub 2024 Jan 9. J Virol. 2024. PMID: 38193694 Free PMC article.
-
Probing Gag-Env dynamics at HIV-1 assembly sites using live-cell microscopy.J Virol. 2024 Sep 17;98(9):e0064924. doi: 10.1128/jvi.00649-24. Epub 2024 Aug 13. J Virol. 2024. PMID: 39136462 Free PMC article.
References
-
- Chertova E, Bess JW Jr, Crise BJ, Sowder IR, Schaden TM, et al. (2002) Envelope glycoprotein incorporation, not shedding of surface envelope glycoprotein (gp120/SU), Is the primary determinant of SU content of purified human immunodeficiency virus type 1 and simian immunodeficiency virus. J Virol 76: 5315–5325. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

