Allele-specific marker development and selection efficiencies for both flavonoid 3'-hydroxylase and flavonoid 3',5'-hydroxylase genes in soybean subgenus soja
- PMID: 23463490
- PMCID: PMC3664743
- DOI: 10.1007/s00122-013-2063-3
Allele-specific marker development and selection efficiencies for both flavonoid 3'-hydroxylase and flavonoid 3',5'-hydroxylase genes in soybean subgenus soja
Abstract
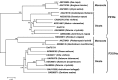
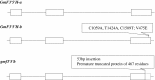
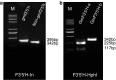
Color is one of the phenotypic markers mostly used to study soybean (Glycine max L. Merr.) genetic, molecular and biochemical processes. Two P450-dependent mono-oxygenases, flavonoid 3'-hydroxylase (F3'H; EC1.14.3.21) and flavonoid 3',5'-hydroxylase (F3'5'H, EC1.14.13.88), both catalyzing the hydroxylation of the B-ring in flavonoids, play an important role in coloration. Previous studies showed that the T locus was a gene encoding F3'H and the W1 locus co-segregated with a gene encoding F3'5'H in soybean. These two genetic loci have identified to control seed coat, flower and pubescence colors. However, the allelic distributions of both F3'H and F3'5'H genes in soybean were unknown. In this study, three novel alleles were identified (two of four alleles for GmF3'H and one of three alleles for GmF3'5'H). A set of gene-tagged markers was developed and verified based on the sequence diversity of all seven alleles. Furthermore, the markers were used to analyze soybean accessions including 170 cultivated soybeans (G. max) from a mini core collection and 102 wild soybeans (G. soja). For both F3'H and F3'5'H, the marker selection efficiencies for pubescence color and flower color were determined. The results showed that one GmF3'H allele explained 92.2 % of the variation in tawny and two gmf3'h alleles explained 63.8 % of the variation in gray pubescence colors. In addition, two GmF3'5'H alleles and one gmF3'5'h allele explained 94.0 % of the variation in purple and 75.3 % in white flowers, respectively. By the combination of the two loci, seed coat color was determined. In total, 90.9 % of accessions possessing both the gmf3'h-b and gmf3'5'h alleles had yellow seed coats. Therefore, seed coat colors are controlled by more than two loci.
Figures


Similar articles
-
A platform for soybean molecular breeding: the utilization of core collections for food security.Plant Mol Biol. 2013 Sep;83(1-2):41-50. doi: 10.1007/s11103-013-0076-6. Epub 2013 May 25. Plant Mol Biol. 2013. PMID: 23708950 Free PMC article. Review.
-
A single-base deletion in soybean flavonoid 3'-hydroxylase gene is associated with gray pubescence color.Plant Mol Biol. 2002 Sep;50(2):187-96. doi: 10.1023/a:1016087221334. Plant Mol Biol. 2002. PMID: 12175012
-
A new allele of flower color gene W1 encoding flavonoid 3'5'-hydroxylase is responsible for light purple flowers in wild soybean Glycine soja.BMC Plant Biol. 2010 Jul 28;10:155. doi: 10.1186/1471-2229-10-155. BMC Plant Biol. 2010. PMID: 20663233 Free PMC article.
-
Cloning of the pleiotropic T locus in soybean and two recessive alleles that differentially affect structure and expression of the encoded flavonoid 3' hydroxylase.Genetics. 2003 Jan;163(1):295-309. doi: 10.1093/genetics/163.1.295. Genetics. 2003. PMID: 12586717 Free PMC article.
-
Flower colour and cytochromes P450.Philos Trans R Soc Lond B Biol Sci. 2013 Jan 6;368(1612):20120432. doi: 10.1098/rstb.2012.0432. Print 2013 Feb 19. Philos Trans R Soc Lond B Biol Sci. 2013. PMID: 23297355 Free PMC article. Review.
Cited by
-
Characterization of the Common Genetic Basis Underlying Seed Hilum Size, Yield, and Quality Traits in Soybean.Front Plant Sci. 2021 Feb 25;12:610214. doi: 10.3389/fpls.2021.610214. eCollection 2021. Front Plant Sci. 2021. PMID: 33719282 Free PMC article.
-
The Effects of Domestication on Secondary Metabolite Composition in Legumes.Front Genet. 2020 Sep 18;11:581357. doi: 10.3389/fgene.2020.581357. eCollection 2020. Front Genet. 2020. PMID: 33193705 Free PMC article. Review.
-
A platform for soybean molecular breeding: the utilization of core collections for food security.Plant Mol Biol. 2013 Sep;83(1-2):41-50. doi: 10.1007/s11103-013-0076-6. Epub 2013 May 25. Plant Mol Biol. 2013. PMID: 23708950 Free PMC article. Review.
-
Identification of candidate genes for soybean seed coat-related traits using QTL mapping and GWAS.Front Plant Sci. 2023 Jun 13;14:1190503. doi: 10.3389/fpls.2023.1190503. eCollection 2023. Front Plant Sci. 2023. PMID: 37384360 Free PMC article.
-
Heterologous Expression of Platycodon grandiflorus PgF3'5'H Modifies Flower Color Pigmentation in Tobacco.Genes (Basel). 2023 Oct 9;14(10):1920. doi: 10.3390/genes14101920. Genes (Basel). 2023. PMID: 37895269 Free PMC article.
References
-
- Cheng H, Yu O, Yu D-Y. Polymorphisms of IFS1 and IFS2 gene are associated with isoflavone concentrations in soybean seeds. Plant Sci. 2008;175:505–512. doi: 10.1016/j.plantsci.2008.05.020. - DOI
-
- de Vetten N, ter Horst J, van Schaik HP, de Boer A, Mol J, Koes R. A cytochrome b5 is required for full activity of flavonoid 3′, 5′-hydroxylase, a cytochrome P450 involved in the formation of blue flower colors. Proc Natl Acad Sci USA. 1999;96:778–783. doi: 10.1073/pnas.96.2.778. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

