Regulation of molecular chaperones through post-translational modifications: decrypting the chaperone code
- PMID: 23459247
- PMCID: PMC4492711
- DOI: 10.1016/j.bbagrm.2013.02.010
Regulation of molecular chaperones through post-translational modifications: decrypting the chaperone code
Abstract
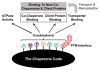
Molecular chaperones and their associated cofactors form a group of highly specialized proteins that orchestrate the folding and unfolding of other proteins and the assembly and disassembly of protein complexes. Chaperones are found in all cell types and organisms, and their activity must be tightly regulated to maintain normal cell function. Indeed, deregulation of protein folding and protein complex assembly is the cause of various human diseases. Here, we present the results of an extensive review of the literature revealing that the post-translational modification (PTM) of chaperones has been selected during evolution as an efficient mean to regulate the activity and specificity of these key proteins. Because the addition and reciprocal removal of chemical groups can be triggered very rapidly, this mechanism provides an efficient switch to precisely regulate the activity of chaperones on specific substrates. The large number of PTMs detected in chaperones suggests that a combinatory code is at play to regulate function, activity, localization, and substrate specificity for this group of biologically important proteins. This review surveys the core information currently available as a starting point toward the more ambitious endeavor of deciphering the "chaperone code".
Copyright © 2013 Elsevier B.V. All rights reserved.
Figures


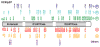
Similar articles
-
Post-translational modifications of Hsp70 family proteins: Expanding the chaperone code.J Biol Chem. 2020 Jul 31;295(31):10689-10708. doi: 10.1074/jbc.REV120.011666. Epub 2020 Jun 9. J Biol Chem. 2020. PMID: 32518165 Free PMC article. Review.
-
Mechanisms targeting apolipoprotein B100 to proteasomal degradation: evidence that degradation is initiated by BiP binding at the N terminus and the formation of a p97 complex at the C terminus.Arterioscler Thromb Vasc Biol. 2009 Apr;29(4):579-85. doi: 10.1161/ATVBAHA.108.181859. Epub 2009 Jan 22. Arterioscler Thromb Vasc Biol. 2009. PMID: 19164805
-
Mitochondrial Chaperone Code: Just warming up.Cell Stress Chaperones. 2024 Jun;29(3):483-496. doi: 10.1016/j.cstres.2024.05.002. Epub 2024 May 17. Cell Stress Chaperones. 2024. PMID: 38763405 Free PMC article. Review.
-
Cdc37-Hsp90 complexes are responsive to nucleotide-induced conformational changes and binding of further cofactors.J Biol Chem. 2010 Dec 24;285(52):40921-32. doi: 10.1074/jbc.M110.131086. Epub 2010 Sep 29. J Biol Chem. 2010. PMID: 20880838 Free PMC article.
-
Molecular chaperones HscA/Ssq1 and HscB/Jac1 and their roles in iron-sulfur protein maturation.Crit Rev Biochem Mol Biol. 2007 Mar-Apr;42(2):95-111. doi: 10.1080/10409230701322298. Crit Rev Biochem Mol Biol. 2007. PMID: 17453917 Review.
Cited by
-
TRAP1 Chaperones the Metabolic Switch in Cancer.Biomolecules. 2022 Jun 4;12(6):786. doi: 10.3390/biom12060786. Biomolecules. 2022. PMID: 35740911 Free PMC article. Review.
-
CD40 in Endothelial Cells Restricts Neural Tissue Invasion by Toxoplasma gondii.Infect Immun. 2019 Jul 23;87(8):e00868-18. doi: 10.1128/IAI.00868-18. Print 2019 Aug. Infect Immun. 2019. PMID: 31109947 Free PMC article.
-
A proteolytic pathway that controls glucose uptake in fat and muscle.Rev Endocr Metab Disord. 2014 Mar;15(1):55-66. doi: 10.1007/s11154-013-9276-2. Rev Endocr Metab Disord. 2014. PMID: 24114239 Free PMC article. Review.
-
Molecular Stressors Engender Protein Connectivity Dysfunction through Aberrant N-Glycosylation of a Chaperone.Cell Rep. 2020 Jun 30;31(13):107840. doi: 10.1016/j.celrep.2020.107840. Cell Rep. 2020. PMID: 32610141 Free PMC article.
-
Identification genetic variations in some heat shock protein genes of Tali goat breed and study their structural and functional effects on relevant proteins.Vet Med Sci. 2023 Sep;9(5):2247-2259. doi: 10.1002/vms3.1231. Epub 2023 Aug 2. Vet Med Sci. 2023. PMID: 37530404 Free PMC article.
References
-
- Tissieres A, Mitchell HK, Tracy UM. Protein synthesis in salivary glands of Drosophila melanogaster: relation to chromosome puffs. J Mol Biol. 1974;84:389–398. - PubMed
-
- Frydman J, Nimmesgern E, Ohtsuka K, Hartl FU. Folding of nascent polypeptide chains in a high molecular mass assembly with molecular chaperones. Nature. 1994;370:111–117. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

