The human kinome and kinase inhibition
- PMID: 23456613
- PMCID: PMC4128285
- DOI: 10.1002/0471141755.ph0209s60
The human kinome and kinase inhibition
Abstract
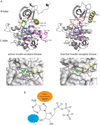
Protein and lipid kinases play key regulatory roles in a number of biological processes. Unsurprisingly, activating mutations in kinases have been linked to a number of disorders and diseases, most notably cancers. Thus, kinases have emerged as promising clinical targets. There are more than 500 human protein kinases and about 20 lipid kinases. Most protein kinases share a highly conserved domain, the eukaryotic protein kinase (ePK) domain, which contains the ATP and substrate-binding sites. Many inhibitors in clinical use bind to the highly conserved ATP binding site. For this reason, many kinase inhibitors are not exclusively selective for their intended targets. Furthermore, despite the current interest in kinase inhibitors, very few kinases implicated in disease have validated inhibitors. This unit describes the human kinome, ePK structure, and types of kinase inhibitors, focusing on methods to identify potent and selective kinase inhibitors.
Curr. Protoc. Pharmacol. 60:2.9.1-2.9.14. © 2013 by John Wiley & Sons, Inc.
Figures


Similar articles
-
Large-scale proteomics analysis of the human kinome.Mol Cell Proteomics. 2009 Jul;8(7):1751-64. doi: 10.1074/mcp.M800588-MCP200. Epub 2009 Apr 15. Mol Cell Proteomics. 2009. PMID: 19369195 Free PMC article.
-
Protein kinase inhibitors: contributions from structure to clinical compounds.Q Rev Biophys. 2009 Feb;42(1):1-40. doi: 10.1017/S0033583508004745. Epub 2009 Mar 19. Q Rev Biophys. 2009. PMID: 19296866 Review.
-
Comprehensive Data-Driven Assessment of Non-Kinase Targets of Inhibitors of the Human Kinome.Biomolecules. 2024 Feb 21;14(3):258. doi: 10.3390/biom14030258. Biomolecules. 2024. PMID: 38540679 Free PMC article. Review.
-
Chemical proteomics and functional proteomics strategies for protein kinase inhibitor validation and protein kinase substrate identification: applications to protein kinase CK2.Biochim Biophys Acta. 2013 Jul;1834(7):1352-8. doi: 10.1016/j.bbapap.2013.02.006. Epub 2013 Feb 14. Biochim Biophys Acta. 2013. PMID: 23416530 Review.
-
The dynamic nature of the kinome.Biochem J. 2013 Feb 15;450(1):1-8. doi: 10.1042/BJ20121456. Biochem J. 2013. PMID: 23343193 Free PMC article. Review.
Cited by
-
Molecular Networking Revealed Unique UV-Absorbing Phospholipids: Favilipids from the Marine Sponge Clathria faviformis.Mar Drugs. 2023 Jan 18;21(2):58. doi: 10.3390/md21020058. Mar Drugs. 2023. PMID: 36827099 Free PMC article.
-
kinCSM: Using graph-based signatures to predict small molecule CDK2 inhibitors.Protein Sci. 2022 Nov;31(11):e4453. doi: 10.1002/pro.4453. Protein Sci. 2022. PMID: 36305769 Free PMC article.
-
Coral: Clear and Customizable Visualization of Human Kinome Data.Cell Syst. 2018 Sep 26;7(3):347-350.e1. doi: 10.1016/j.cels.2018.07.001. Epub 2018 Aug 29. Cell Syst. 2018. PMID: 30172842 Free PMC article.
-
PKMYT1 Is a Marker of Treatment Response and a Therapeutic Target for CDK4/6 Inhibitor-Resistance in ER+ Breast Cancer.Mol Cancer Ther. 2024 Oct 1;23(10):1494-1510. doi: 10.1158/1535-7163.MCT-23-0564. Mol Cancer Ther. 2024. PMID: 38781103 Free PMC article.
-
Synthetic Heterocyclic Derivatives as Kinase Inhibitors Tested for the Treatment of Neuroblastoma.Molecules. 2021 Nov 23;26(23):7069. doi: 10.3390/molecules26237069. Molecules. 2021. PMID: 34885651 Free PMC article. Review.
References
-
- Adrian FJ, Ding Q, Sim T, Velentza A, Sloan C, Liu Y, Zhang G, Hur W, Ding S, Manley P, Mestan J, Fabbro D, Gray NS. Allosteric inhibitors of Bcr-abl-dependent cell proliferation. Nat. Chem. Biol. 2006;2:95–102. - PubMed
-
- Barf T, Kaptein A. Irreversible protein kinase inhibitors: balancing the benefits and risks. J. Med. Chem. 2012;55:6243–6262. - PubMed
-
- Ben-Neriah Y, Daley GQ, Mes-Masson AM, Witte ON, Baltimore D. The chronic myelogenous leukemia-specific P210 protein is the product of the bcr/abl hybrid gene. Science. 1986;233:212–214. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

