Whole-exome sequencing identifies mutated c12orf57 in recessive corpus callosum hypoplasia
- PMID: 23453666
- PMCID: PMC3591854
- DOI: 10.1016/j.ajhg.2013.02.004
Whole-exome sequencing identifies mutated c12orf57 in recessive corpus callosum hypoplasia
Abstract
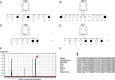
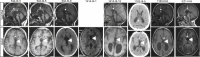
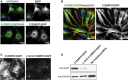
The corpus callosum is the principal cerebral commissure connecting the right and left hemispheres. The development of the corpus callosum is under tight genetic control, as demonstrated by abnormalities in its development in more than 1,000 genetic syndromes. We recruited more than 25 families in which members affected with corpus callosum hypoplasia (CCH) lacked syndromic features and had consanguineous parents, suggesting recessive causes. Exome sequence analysis identified C12orf57 mutations at the initiator methionine codon in four different families. C12orf57 is ubiquitously expressed and encodes a poorly annotated 126 amino acid protein of unknown function. This protein is without significant paralogs but has been tightly conserved across evolution. Our data suggest that this conserved gene is required for development of the human corpus callosum.
Copyright © 2013 The American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures

Similar articles
-
Exome sequencing identifies compound heterozygous mutations in C12orf57 in two siblings with severe intellectual disability, hypoplasia of the corpus callosum, chorioretinal coloboma, and intractable seizures.Am J Med Genet A. 2014 Aug;164A(8):1976-80. doi: 10.1002/ajmg.a.36592. Epub 2014 May 5. Am J Med Genet A. 2014. PMID: 24798461 Review.
-
Novel ABCD1 Gene Mutation in Adrenomyeloneuropathy with Hypoplasia and Agenesis of the Corpus Callosum.Neurodegener Dis. 2018;18(2-3):156-164. doi: 10.1159/000490248. Epub 2018 Jul 2. Neurodegener Dis. 2018. PMID: 29966135
-
Clinical, genetic and imaging findings identify new causes for corpus callosum development syndromes.Brain. 2014 Jun;137(Pt 6):1579-613. doi: 10.1093/brain/awt358. Epub 2014 Jan 28. Brain. 2014. PMID: 24477430 Free PMC article. Review.
-
Exome sequencing identifies recessive CDK5RAP2 variants in patients with isolated agenesis of corpus callosum.Eur J Hum Genet. 2016 Apr;24(4):607-10. doi: 10.1038/ejhg.2015.156. Epub 2015 Jul 22. Eur J Hum Genet. 2016. PMID: 26197979 Free PMC article.
-
Further delineation of Temtamy syndrome of corpus callosum and ocular abnormalities.Am J Med Genet A. 2018 Mar;176(3):715-721. doi: 10.1002/ajmg.a.38615. Epub 2018 Jan 31. Am J Med Genet A. 2018. PMID: 29383837
Cited by
-
Whole exome analysis identifies dominant COL4A1 mutations in patients with complex ocular phenotypes involving microphthalmia.Clin Genet. 2014 Nov;86(5):475-81. doi: 10.1111/cge.12379. Epub 2014 Apr 12. Clin Genet. 2014. PMID: 24628545 Free PMC article.
-
Pre- and Postnatal Analysis of Chromosome 1q44 Deletion in Agenesis of Corpus Callosum.Mol Syndromol. 2015 Oct;6(4):187-92. doi: 10.1159/000440659. Epub 2015 Sep 11. Mol Syndromol. 2015. PMID: 26648835 Free PMC article.
-
The Impact of the IKBKG Gene on the Appearance of the Corpus Callosum Abnormalities in Incontinentia Pigmenti.Diagnostics (Basel). 2023 Mar 30;13(7):1300. doi: 10.3390/diagnostics13071300. Diagnostics (Basel). 2023. PMID: 37046518 Free PMC article.
-
Genomic variants and variations in malformations of cortical development.Pediatr Clin North Am. 2015 Jun;62(3):571-85. doi: 10.1016/j.pcl.2015.03.002. Epub 2015 Apr 1. Pediatr Clin North Am. 2015. PMID: 26022163 Free PMC article. Review.
-
Genes with high penetrance for syndromic and non-syndromic autism typically function within the nucleus and regulate gene expression.Mol Autism. 2016 Mar 15;7:18. doi: 10.1186/s13229-016-0082-z. eCollection 2016. Mol Autism. 2016. PMID: 26985359 Free PMC article.
References
-
- Scamvougeras A., Kigar D.L., Jones D., Weinberger D.R., Witelson S.F. Size of the human corpus callosum is genetically determined: an MRI study in mono and dizygotic twins. Neurosci. Lett. 2003;338:91–94. - PubMed
-
- Sztriha L. Spectrum of corpus callosum agenesis. Pediatr. Neurol. 2005;32:94–101. - PubMed
-
- Murray S.S., Oliphant A., Shen R., McBride C., Steeke R.J., Shannon S.G., Rubano T., Kermani B.G., Fan J.B., Chee M.S., Hansen M.S. A highly informative SNP linkage panel for human genetic studies. Nat. Methods. 2004;1:113–117. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

