The nonstructural protein 2C of a Picorna-like virus displays nucleic acid helix destabilizing activity that can be functionally separated from its ATPase activity
- PMID: 23449794
- PMCID: PMC3624285
- DOI: 10.1128/JVI.00245-13
The nonstructural protein 2C of a Picorna-like virus displays nucleic acid helix destabilizing activity that can be functionally separated from its ATPase activity
Abstract
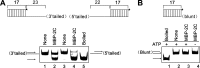
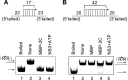
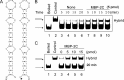
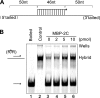
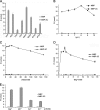
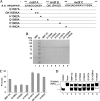
Picorna-like viruses in the Picornavirales order are a large group of positive-strand RNA viruses that include numerous important pathogens for plants, insects, and humans. In these viruses, nonstructural protein 2C is one of the most conserved proteins and contains ATPase activity and putative RNA helicase activity. Here we expressed 2C protein of Ectropis obliqua picorna-like virus (EoV; genus Iflavirus, family Iflaviridae, order Picornavirales) in a eukaryotic expression system and determined that EoV 2C displays ATP-independent nucleic acid helix destabilizing and strand annealing acceleration activity in a concentration-dependent manner, indicating that this picornaviral 2C is more like an RNA chaperone than like the previously predicted RNA helicase. Our further characterization of EoV 2C revealed that divalent metal ions, such as Mg(2+) and Zn(2+), inhibit 2C-mediated helix destabilization to different extents. Moreover, we determined that EoV 2C also contains ATPase activity like that of other picornaviral 2C proteins and further assessed the functional relevance between its RNA chaperone-like and ATPase activities using mutational analysis as well as their responses to Mg(2+). Our data show that, when one of the two 2C activities was dramatically inhibited or almost abolished, the other activity could remain intact, showing that the RNA chaperone-like and ATPase activities of EoV 2C can be functionally separated. This report reveals that a picorna-like virus 2C protein displays RNA helix destabilizing and strand annealing acceleration activity, which may be critical for picornaviral replication and pathogenesis, and should foster our understanding of picorna-like viruses and viral RNA chaperones.
Figures





Similar articles
-
The identification and characterization of nucleic acid chaperone activity of human enterovirus 71 nonstructural protein 3AB.Virology. 2014 Sep;464-465:353-364. doi: 10.1016/j.virol.2014.07.037. Epub 2014 Aug 9. Virology. 2014. PMID: 25113906 Free PMC article.
-
Sequence analysis and genomic organization of a new insect picorna-like virus, Ectropis obliqua picorna-like virus, isolated from Ectropis obliqua.J Gen Virol. 2004 May;85(Pt 5):1145-1151. doi: 10.1099/vir.0.19638-0. J Gen Virol. 2004. PMID: 15105531
-
Human Norovirus NS3 Has RNA Helicase and Chaperoning Activities.J Virol. 2018 Feb 12;92(5):e01606-17. doi: 10.1128/JVI.01606-17. Print 2018 Mar 1. J Virol. 2018. PMID: 29237842 Free PMC article.
-
Evolution and taxonomy of positive-strand RNA viruses: implications of comparative analysis of amino acid sequences.Crit Rev Biochem Mol Biol. 1993;28(5):375-430. doi: 10.3109/10409239309078440. Crit Rev Biochem Mol Biol. 1993. PMID: 8269709 Review.
-
RNA chaperones encoded by RNA viruses.Virol Sin. 2015 Dec;30(6):401-9. doi: 10.1007/s12250-015-3676-2. Epub 2015 Dec 22. Virol Sin. 2015. PMID: 26715301 Free PMC article. Review.
Cited by
-
A Novel Iflavirus Was Discovered in Green Rice Leafhopper Nephotettix cincticeps and Its Proliferation Was Inhibited by Infection of Rice Dwarf Virus.Front Microbiol. 2021 Jan 8;11:621141. doi: 10.3389/fmicb.2020.621141. eCollection 2020. Front Microbiol. 2021. PMID: 33488564 Free PMC article.
-
The identification and characterization of nucleic acid chaperone activity of human enterovirus 71 nonstructural protein 3AB.Virology. 2014 Sep;464-465:353-364. doi: 10.1016/j.virol.2014.07.037. Epub 2014 Aug 9. Virology. 2014. PMID: 25113906 Free PMC article.
-
Fluvoxamine: A Review of Its Mechanism of Action and Its Role in COVID-19.Front Pharmacol. 2021 Apr 20;12:652688. doi: 10.3389/fphar.2021.652688. eCollection 2021. Front Pharmacol. 2021. PMID: 33959018 Free PMC article.
-
Antiviral Peptides Targeting the Helicase Activity of Enterovirus Nonstructural Protein 2C.J Virol. 2021 May 24;95(12):e02324-20. doi: 10.1128/JVI.02324-20. Print 2021 May 24. J Virol. 2021. PMID: 33789997 Free PMC article.
-
Ebola virus VP35 has novel NTPase and helicase-like activities.Nucleic Acids Res. 2019 Jun 20;47(11):5837-5851. doi: 10.1093/nar/gkz340. Nucleic Acids Res. 2019. PMID: 31066445 Free PMC article.
References
-
- Koonin EV, Wolf YI, Nagasaki K, Dolja VV. 2008. The Big Bang of picorna-like virus evolution antedates the radiation of eukaryotic supergroups. Nat. Rev. Microbiol. 6:925–939 - PubMed
-
- Kew O, Morris-Glasgow V, Landaverde M, Burns C, Shaw J, Garib Z, Andre J, Blackman E, Freeman CJ, Jorba J, Sutter R, Tambini G, Venczel L, Pedreira C, Laender F, Shimizu H, Yoneyama T, Miyamura T, van Der Avoort H, Oberste MS, Kilpatrick D, Cochi S, Pallansch M, de Quadros C. 2002. Outbreak of poliomyelitis in Hispaniola associated with circulating type 1 vaccine-derived poliovirus. Science 296:356–359 - PubMed
-
- Wong SS, Yip CC, Lau SK, Yuen KY. 2010. Human enterovirus 71 and hand, foot and mouth disease. Epidemiol. Infect. 138:1071–1089 - PubMed
-
- Brundage SC, Fitzpatrick AN. 2006. Hepatitis A. Am. Fam. Physician 73:2162–2168 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

