Arp2/3 complex ATP hydrolysis promotes lamellipodial actin network disassembly but is dispensable for assembly
- PMID: 23439681
- PMCID: PMC3587832
- DOI: 10.1083/jcb.201211069
Arp2/3 complex ATP hydrolysis promotes lamellipodial actin network disassembly but is dispensable for assembly
Abstract
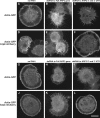
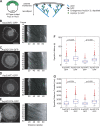
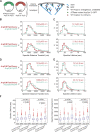
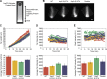
We examined the role of ATP hydrolysis by the Arp2/3 complex in building the leading edge of a cell by studying the effects of hydrolysis defects on the behavior of the complex in the lamellipodial actin network of Drosophila S2 cells and in a reconstituted, in vitro, actin-based motility system. In S2 cells, nonhydrolyzing Arp2 and Arp3 subunits expanded and delayed disassembly of lamellipodial actin networks and the effect of mutant subunits was additive. Arp2 and Arp3 ATP hydrolysis mutants remained in lamellipodial networks longer and traveled greater distances from the plasma membrane, even in networks still containing wild-type Arp2/3 complex. In vitro, wild-type and ATP hydrolysis mutant Arp2/3 complexes each nucleated actin and built similar dendritic networks. However, networks constructed with Arp2/3 hydrolysis-defective mutants were more resistant to disassembly by cofilin. Our results indicate that ATP hydrolysis on both Arp2 and Arp3 contributes to dissociation of the complex from the actin network but is not strictly necessary for lamellipodial network disassembly.
Figures



Similar articles
-
Arp2/3 ATP hydrolysis-catalysed branch dissociation is critical for endocytic force generation.Nat Cell Biol. 2006 Aug;8(8):826-33. doi: 10.1038/ncb1443. Epub 2006 Jul 23. Nat Cell Biol. 2006. PMID: 16862144
-
Identification of an ATP-controlled allosteric switch that controls actin filament nucleation by Arp2/3 complex.Nat Commun. 2016 Jul 15;7:12226. doi: 10.1038/ncomms12226. Nat Commun. 2016. PMID: 27417392 Free PMC article.
-
Spatial and temporal relationships between actin-filament nucleation, capping, and disassembly.Curr Biol. 2007 Mar 6;17(5):395-406. doi: 10.1016/j.cub.2007.02.012. Curr Biol. 2007. PMID: 17331727 Free PMC article.
-
Control of actin filament treadmilling in cell motility.Annu Rev Biophys. 2010;39:449-70. doi: 10.1146/annurev-biophys-051309-103849. Annu Rev Biophys. 2010. PMID: 20192778 Review.
-
Actin-based motility as a self-organized system: mechanism and reconstitution in vitro.C R Biol. 2003 Feb;326(2):161-70. doi: 10.1016/s1631-0691(03)00067-2. C R Biol. 2003. PMID: 12754935 Review.
Cited by
-
Internetwork competition for monomers governs actin cytoskeleton organization.Nat Rev Mol Cell Biol. 2016 Dec;17(12):799-810. doi: 10.1038/nrm.2016.106. Epub 2016 Sep 14. Nat Rev Mol Cell Biol. 2016. PMID: 27625321 Free PMC article.
-
Force and phosphate release from Arp2/3 complex promote dissociation of actin filament branches.Proc Natl Acad Sci U S A. 2020 Jun 16;117(24):13519-13528. doi: 10.1073/pnas.1911183117. Epub 2020 May 27. Proc Natl Acad Sci U S A. 2020. PMID: 32461373 Free PMC article.
-
Transition State of Arp2/3 Complex Activation by Actin-Bound Dimeric Nucleation-Promoting Factor.Proc Natl Acad Sci U S A. 2023 Aug 15;120(33):e2306165120. doi: 10.1073/pnas.2306165120. Epub 2023 Aug 7. Proc Natl Acad Sci U S A. 2023. PMID: 37549294 Free PMC article.
-
Structural basis for regulation of Arp2/3 complex by GMF.Nat Struct Mol Biol. 2013 Sep;20(9):1062-8. doi: 10.1038/nsmb.2628. Epub 2013 Jul 28. Nat Struct Mol Biol. 2013. PMID: 23893131 Free PMC article.
-
Nampt-mediated spindle sizing secures a post-anaphase increase in spindle speed required for extreme asymmetry.Nat Commun. 2020 Jul 7;11(1):3393. doi: 10.1038/s41467-020-17088-6. Nat Commun. 2020. PMID: 32636388 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

