Computational analysis of KRAS mutations: implications for different effects on the KRAS p.G12D and p.G13D mutations
- PMID: 23437064
- PMCID: PMC3577811
- DOI: 10.1371/journal.pone.0055793
Computational analysis of KRAS mutations: implications for different effects on the KRAS p.G12D and p.G13D mutations
Abstract
Background: The issue of whether patients diagnosed with metastatic colorectal cancer who harbor KRAS codon 13 mutations could benefit from the addition of anti-epidermal growth factor receptor therapy remains under debate. The aim of the current study was to perform computational analysis to investigate the structural implications of the underlying mutations caused by c.38G>A (p.G13D) on protein conformation.
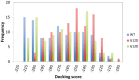
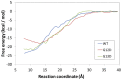
Methods: Molecular dynamics (MD) simulations were performed to understand the plausible structural and dynamical implications caused by c.35G>A (p.G12D) and c.38G>A (p.G13D). The potential of mean force (PMF) simulations were carried out to determine the free energy profiles of the binding processes of GTP interacting with wild-type (WT) KRAS and its mutants (MT).
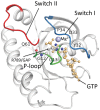
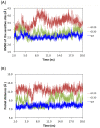
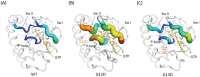
Results: Using MD simulations, we observed that the root mean square deviation (RMSD) increased as a function of time for the MT c.35G>A (p.G12D) and MT c.38G>A (p.G13D) when compared with the WT. We also observed that the GTP-binding pocket in the c.35G>A (p.G12D) mutant is more open than that of the WT and the c.38G>A (p.G13D) proteins. Intriguingly, the analysis of atomic fluctuations and free energy profiles revealed that the mutation of c.35G>A (p.G12D) may induce additional fluctuations in the sensitive sites (P-loop, switch I and II regions). Such fluctuations may promote instability in these protein regions and hamper GTP binding.
Conclusions: Taken together with the results obtained from MD and PMF simulations, the present findings implicate fluctuations at the sensitive sites (P-loop, switch I and II regions). Our findings revealed that KRAS mutations in codon 13 have similar behavior as KRAS WT. To gain a better insight into why patients with metastatic colorectal cancer (mCRC) and the KRAS c.38G>A (p.G13D) mutation appear to benefit from anti-EGFR therapy, the role of the KRAS c.38G>A (p.G13D) mutation in mCRC needs to be further investigated.
Conflict of interest statement
Figures
Similar articles
-
KRAS p.G13D mutation and codon 12 mutations are not created equal in predicting clinical outcomes of cetuximab in metastatic colorectal cancer: a systematic review and meta-analysis.Cancer. 2013 Feb 15;119(4):714-21. doi: 10.1002/cncr.27804. Epub 2012 Sep 12. Cancer. 2013. PMID: 22972628 Review.
-
Meta-analysis comparing the efficacy of anti-EGFR monoclonal antibody therapy between KRAS G13D and other KRAS mutant metastatic colorectal cancer tumours.Eur J Cancer. 2016 Mar;55:122-30. doi: 10.1016/j.ejca.2015.11.025. Epub 2016 Jan 23. Eur J Cancer. 2016. PMID: 26812186 Review.
-
Association of KRAS p.G13D mutation with outcome in patients with chemotherapy-refractory metastatic colorectal cancer treated with cetuximab.JAMA. 2010 Oct 27;304(16):1812-20. doi: 10.1001/jama.2010.1535. JAMA. 2010. PMID: 20978259
-
Impact of genetic profiles on the efficacy of anti-EGFR antibodies in metastatic colorectal cancer with KRAS mutation.Oncol Rep. 2014 Jul;32(1):57-64. doi: 10.3892/or.2014.3179. Epub 2014 May 15. Oncol Rep. 2014. PMID: 24839940
-
Mutation of KRAS in colorectal adenocarcinoma in Greenland.APMIS. 2022 Aug;130(8):493-497. doi: 10.1111/apm.13254. Epub 2022 Jun 20. APMIS. 2022. PMID: 35655437
Cited by
-
Progress and opportunities in molecular pathological epidemiology of colorectal premalignant lesions.Am J Gastroenterol. 2014 Aug;109(8):1205-14. doi: 10.1038/ajg.2014.153. Epub 2014 Jun 17. Am J Gastroenterol. 2014. PMID: 24935274 Free PMC article. Review.
-
Colorectal carcinomas with KRAS codon 12 mutation are associated with more advanced tumor stages.BMC Cancer. 2015 May 1;15:340. doi: 10.1186/s12885-015-1345-3. BMC Cancer. 2015. PMID: 25929517 Free PMC article.
-
A specific KRAS codon 13 mutation is an independent predictor for colorectal cancer metachronous distant metastases.Am J Cancer Res. 2015 Jan 15;5(2):674-88. eCollection 2015. Am J Cancer Res. 2015. PMID: 25973306 Free PMC article.
-
Studying early structural changes in SOS1 mediated KRAS activation mechanism.Curr Res Struct Biol. 2023 Dec 9;7:100115. doi: 10.1016/j.crstbi.2023.100115. eCollection 2024. Curr Res Struct Biol. 2023. PMID: 38188543 Free PMC article.
-
Modeling RAS phenotype in colorectal cancer uncovers novel molecular traits of RAS dependency and improves prediction of response to targeted agents in patients.Clin Cancer Res. 2014 Jan 1;20(1):265-272. doi: 10.1158/1078-0432.CCR-13-1943. Epub 2013 Oct 29. Clin Cancer Res. 2014. PMID: 24170544 Free PMC article.
References
-
- Jemal A, Bray F (2011) Center MM, Ferlay J, Ward E, et al (2011) Global cancer statistics. CA: a cancer journal for clinicians 61: 69–90. - PubMed
-
- American Cancer Society. Cancer Facts & Figures 2009. Available: http://www.cancer.org/Research/CancerFactsFigures/CancerFactsFigures/. Accessed 2013 Jan 7.
-
- (2004) US Food and Drug Administration. FDA approves Erbitux for colorectal cancer. Www Fda Gov.
-
- (2007) US Food and Drug Administration. FDA approves Vectibix (panitumumab) to treat metastatic colorectal carcinoma. Www Fda Gov.
-
- KRAS mutations and epidermal growth factor receptor inhibitor therapy in metastatic colorectal cancer. Technol Eval Cent Assess Program Exec Summ 23: 1–3. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

