Affinity capture and identification of host cell factors associated with hepatitis C virus (+) strand subgenomic RNA
- PMID: 23429521
- PMCID: PMC3675812
- DOI: 10.1074/mcp.M112.017020
Affinity capture and identification of host cell factors associated with hepatitis C virus (+) strand subgenomic RNA
Abstract
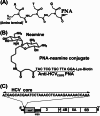
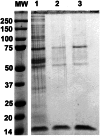
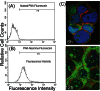
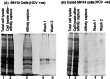
Hepatitis C virus (HCV) infection leading to chronic hepatitis is a major factor in the causation of liver cirrhosis, hepatocellular carcinoma, and liver failure. This process may involve the interplay of various host cell factors, as well as the interaction of these factors with viral RNA and proteins. We report a novel strategy using a sequence-specific biotinylated peptide nucleic acid (PNA)-neamine conjugate targeted to HCV RNA for the in situ capture of subgenomic HCV (+) RNA, along with cellular and viral factors associated with it in MH14 host cells. Using this affinity capture system in conjunction with LC/MS/MS, we have identified 83 cellular factors and three viral proteins (NS5B, NS5A, and NS3-4a protease-helicase) associated with the viral genome. The capture was highly specific. These proteins were not scored with cured MH14 cells devoid of HCV replicons because of the absence of the target sequence in cells for the PNA-neamine probe and also because, unlike oligomeric DNA, cellular proteins have no affinity for PNA. The identified cellular factors belong to different functional groups, including signaling, oncogenic, chaperonin, transcriptional regulators, and RNA helicases as well as DEAD box proteins, ribosomal proteins, translational regulators/factors, and metabolic enzymes, that represent a diverse set of cellular factors associated with the HCV RNA genome. Small interfering RNA-mediated silencing of a diverse class of selected proteins in an HCV replicon cell line either enhanced or inhibited HCV replication/translation, suggesting that these cellular factors have regulatory roles in HCV replication.
Figures


Similar articles
-
Affinity Purification of the Hepatitis C Virus Replicase Identifies Valosin-Containing Protein, a Member of the ATPases Associated with Diverse Cellular Activities Family, as an Active Virus Replication Modulator.J Virol. 2016 Oct 14;90(21):9953-9966. doi: 10.1128/JVI.01140-16. Print 2016 Nov 1. J Virol. 2016. PMID: 27558430 Free PMC article.
-
Replicons of a Rodent Hepatitis C Model Virus Permit Selection of Highly Permissive Cells.J Virol. 2019 Sep 12;93(19):e00733-19. doi: 10.1128/JVI.00733-19. Print 2019 Oct 1. J Virol. 2019. PMID: 31292246 Free PMC article.
-
Seed sequence-matched controls reveal limitations of small interfering RNA knockdown in functional and structural studies of hepatitis C virus NS5A-MOBKL1B interaction.J Virol. 2014 Oct;88(19):11022-33. doi: 10.1128/JVI.01582-14. Epub 2014 Jul 16. J Virol. 2014. PMID: 25031347 Free PMC article.
-
Recent advances in the molecular biology of hepatitis C virus.J Mol Biol. 2001 Oct 26;313(3):451-64. doi: 10.1006/jmbi.2001.5055. J Mol Biol. 2001. PMID: 11676530 Review.
-
Hepatitis C virus: virology and life cycle.Clin Mol Hepatol. 2013 Mar;19(1):17-25. doi: 10.3350/cmh.2013.19.1.17. Epub 2013 Mar 25. Clin Mol Hepatol. 2013. PMID: 23593605 Free PMC article. Review.
Cited by
-
Proteomic approaches to analyzing hepatitis C virus biology.Proteomics. 2015 Jun;15(12):2051-65. doi: 10.1002/pmic.201500009. Epub 2015 May 5. Proteomics. 2015. PMID: 25809442 Free PMC article. Review.
-
A new stochastic model for subgenomic hepatitis C virus replication considers drug resistant mutants.PLoS One. 2014 Mar 18;9(3):e91502. doi: 10.1371/journal.pone.0091502. eCollection 2014. PLoS One. 2014. PMID: 24643004 Free PMC article.
-
Effect of P-body component Mov10 on HCV virus production and infectivity.FASEB J. 2020 Jul;34(7):9433-9449. doi: 10.1096/fj.201800641R. Epub 2020 Jun 4. FASEB J. 2020. PMID: 32496609 Free PMC article.
-
RNA-Centric Approaches to Profile the RNA-Protein Interaction Landscape on Selected RNAs.Noncoding RNA. 2021 Feb 15;7(1):11. doi: 10.3390/ncrna7010011. Noncoding RNA. 2021. PMID: 33671874 Free PMC article. Review.
-
Identification of HNRNPK as regulator of hepatitis C virus particle production.PLoS Pathog. 2015 Jan 8;11(1):e1004573. doi: 10.1371/journal.ppat.1004573. eCollection 2015 Jan. PLoS Pathog. 2015. PMID: 25569684 Free PMC article.
References
-
- Rijnbrand R. C., Lemon S. M. (2000) Internal ribosome entry site-mediated translation in hepatitis C virus replication. Curr. Top. Microbiol. Immunol. 242, 85–116 - PubMed
-
- Reed K. E., Rice C. M. (2000) Overview of hepatitis C virus genome structure, polyprotein processing, and protein properties. Curr. Top. Microbiol. Immunol. 242, 55–84 - PubMed
-
- Oh J. W., Sheu G. T., Lai M. M. (2000) Template requirement and initiation site selection by hepatitis C virus polymerase on a minimal viral RNA template. J. Biol. Chem. 275, 17710–17717 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

