PBOV1 is a human de novo gene with tumor-specific expression that is associated with a positive clinical outcome of cancer
- PMID: 23418531
- PMCID: PMC3572036
- DOI: 10.1371/journal.pone.0056162
PBOV1 is a human de novo gene with tumor-specific expression that is associated with a positive clinical outcome of cancer
Abstract
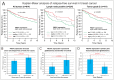
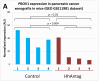
PBOV1 is a known human protein-coding gene with an uncharacterized function. We have previously found that PBOV1 lacks orthologs in non-primate genomes and is expressed in a wide range of tumor types. Here we report that PBOV1 protein-coding sequence is human-specific and has originated de novo in the primate evolution through a series of frame-shift and stop codon mutations. We profiled PBOV1 expression in multiple cancer and normal tissue samples and found that it was expressed in 19 out of 34 tumors of various origins but completely lacked expression in any of the normal adult or fetal human tissues. We found that, unlike the cancer/testis antigens that are typically controlled by CpG island-containing promoters, PBOV1 was expressed from a GC-poor TATA-containing promoter which was not influenced by CpG demethylation and was inactive in testis. Our analysis of public microarray data suggests that PBOV1 activation in tumors could be dependent on the Hedgehog signaling pathway. Despite the recent de novo origin and the lack of identifiable functional signatures, a missense SNP in the PBOV1 coding sequence has been previously associated with an increased risk of breast cancer. Using publicly available microarray datasets, we found that high levels of PBOV1 expression in breast cancer and glioma samples were significantly associated with a positive outcome of the cancer disease. We also found that PBOV1 was highly expressed in primary but not in recurrent high-grade gliomas, suggesting the presence of a negative selection against PBOV1-expressing cancer cells. Our findings could contribute to the understanding of the mechanisms behind de novo gene origin and the possible role of tumors in this process.
Conflict of interest statement
Figures



Similar articles
-
PBOV1 correlates with progression of ovarian cancer and inhibits proliferation of ovarian cancer cells.Oncol Rep. 2016 Jan;35(1):488-96. doi: 10.3892/or.2015.4396. Epub 2015 Nov 4. Oncol Rep. 2016. PMID: 26549570
-
PBOV1 as a potential biomarker for more advanced prostate cancer based on protein and digital histomorphometric analysis.Prostate. 2018 May;78(7):547-559. doi: 10.1002/pros.23499. Epub 2018 Mar 9. Prostate. 2018. PMID: 29520928 Free PMC article.
-
Association between the overexpression of PBOV1 and the prognosis of patients with hepatocellular carcinoma.Oncol Lett. 2018 Sep;16(3):3401-3407. doi: 10.3892/ol.2018.9013. Epub 2018 Jun 25. Oncol Lett. 2018. PMID: 30127941 Free PMC article.
-
[Transcriptomic regulation and molecular mechanism of polygenic tumor at different stages].Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2011 Jul;36(7):585-91. doi: 10.3969/j.issn.1672-7347.2011.07.001. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2011. PMID: 21873780 Review. Chinese.
-
The Origins and Functions of De Novo Genes: Against All Odds?J Mol Evol. 2022 Aug;90(3-4):244-257. doi: 10.1007/s00239-022-10055-3. Epub 2022 Apr 22. J Mol Evol. 2022. PMID: 35451603 Free PMC article. Review.
Cited by
-
New genes contribute to genetic and phenotypic novelties in human evolution.Curr Opin Genet Dev. 2014 Dec;29:90-6. doi: 10.1016/j.gde.2014.08.013. Epub 2014 Sep 16. Curr Opin Genet Dev. 2014. PMID: 25218862 Free PMC article. Review.
-
The Theory of Carcino-Evo-Devo and Its Non-Trivial Predictions.Genes (Basel). 2022 Dec 12;13(12):2347. doi: 10.3390/genes13122347. Genes (Basel). 2022. PMID: 36553613 Free PMC article.
-
Oncogenes, tumor suppressor and differentiation genes represent the oldest human gene classes and evolve concurrently.Sci Rep. 2019 Nov 11;9(1):16410. doi: 10.1038/s41598-019-52835-w. Sci Rep. 2019. PMID: 31712655 Free PMC article.
-
Carcino-Evo-Devo, A Theory of the Evolutionary Role of Hereditary Tumors.Int J Mol Sci. 2023 May 11;24(10):8611. doi: 10.3390/ijms24108611. Int J Mol Sci. 2023. PMID: 37239953 Free PMC article. Review.
-
miR-431-5p alters the epithelial-to-mesenchymal transition markers by targeting UROC28 in hepatoma cells.Onco Targets Ther. 2018 Oct 4;11:6489-6503. doi: 10.2147/OTT.S173840. eCollection 2018. Onco Targets Ther. 2018. PMID: 30323624 Free PMC article.
References
-
- Long M, Betrán E, Thornton K, Wang W (2003) The origin of new genes: glimpses from the young and old. Nature reviews Genetics 4: 865–875. - PubMed
-
- Snel B, Bork P, Huynen MA (2002) Genomes in flux: the evolution of archaeal and proteobacterial gene content. Genome research 12: 17–25. - PubMed
-
- Toll-Riera M, Bosch N, Bellora N, Castelo R, Armengol L, et al. (2009) Origin of primate orphan genes: a comparative genomics approach. Molecular biology and evolution 26: 603–612. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

