A two-component system (XydS/R) controls the expression of genes encoding CBM6-containing proteins in response to straw in Clostridium cellulolyticum
- PMID: 23418511
- PMCID: PMC3572039
- DOI: 10.1371/journal.pone.0056063
A two-component system (XydS/R) controls the expression of genes encoding CBM6-containing proteins in response to straw in Clostridium cellulolyticum
Abstract
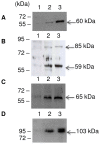
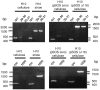
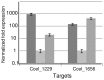
The composition of the cellulosomes (multi enzymatic complexes involved in the degradation of plant cell wall polysaccharides) produced by Clostridium cellulolyticum differs according to the growth substrate. In particular, the expression of a cluster of 14 hemicellulase-encoding genes (called xyl-doc) seems to be induced by the presence of straw and not of cellulose. Genes encoding a putative two-component regulation system (XydS/R) were found upstream of xyl-doc. First evidence for the involvement of the response regulator, XydR, part of this two-component system, in the expression of xyl-doc genes was given by the analysis of the cellulosomes produced by a regulator overproducing strain when grown on cellulose. Nano-LC MS/MS analysis allowed the detection of the products of all xyl-doc genes and of the product of the gene at locus Ccel_1656 predicted to bear a carbohydrate binding domain targeting hemicellulose. RT-PCR experiments further demonstrated that the regulation occurs at the transcriptional level and that all xyl-doc genes are transcriptionally linked. mRNA quantification in a regulator knock-out strain and in its complemented derivative confirmed the involvement of the regulator in the expression of xyl-doc genes and of the gene at locus Ccel_1656 in response to straw. Electrophoretic mobility shift assays using the purified regulator further demonstrated that the regulator binds to DNA regions located upstream of the first gene of the xyl-doc gene cluster and upstream of the gene at locus Ccel_1656.
Conflict of interest statement
Figures


Similar articles
-
Modulation of cellulosome composition in Clostridium cellulolyticum: adaptation to the polysaccharide environment revealed by proteomic and carbohydrate-active enzyme analyses.Proteomics. 2010 Feb;10(3):541-54. doi: 10.1002/pmic.200900311. Proteomics. 2010. PMID: 20013800
-
Are cellulosome scaffolding protein CipC and CBM3-containing protein HycP, involved in adherence of Clostridium cellulolyticum to cellulose?PLoS One. 2013 Jul 25;8(7):e69360. doi: 10.1371/journal.pone.0069360. Print 2013. PLoS One. 2013. PMID: 23935995 Free PMC article.
-
The cellulosomes from Clostridium cellulolyticum: identification of new components and synergies between complexes.FEBS J. 2009 Jun;276(11):3076-86. doi: 10.1111/j.1742-4658.2009.07025.x. Epub 2009 Apr 22. FEBS J. 2009. PMID: 19490109
-
The cellulosomes: multienzyme machines for degradation of plant cell wall polysaccharides.Annu Rev Microbiol. 2004;58:521-54. doi: 10.1146/annurev.micro.57.030502.091022. Annu Rev Microbiol. 2004. PMID: 15487947 Review.
-
Clostridium cellulolyticum: model organism of mesophilic cellulolytic clostridia.FEMS Microbiol Rev. 2005 Sep;29(4):741-64. doi: 10.1016/j.femsre.2004.11.003. Epub 2004 Dec 1. FEMS Microbiol Rev. 2005. PMID: 16102601 Review.
Cited by
-
Elucidation of the recognition mechanisms for hemicellulose and pectin in Clostridium cellulovorans using intracellular quantitative proteome analysis.AMB Express. 2015 May 23;5:29. doi: 10.1186/s13568-015-0115-6. eCollection 2015. AMB Express. 2015. PMID: 26020016 Free PMC article.
-
In vitro and in vivo exploration of the cellobiose and cellodextrin phosphorylases panel in Ruminiclostridium cellulolyticum: implication for cellulose catabolism.Biotechnol Biofuels. 2019 Sep 3;12:208. doi: 10.1186/s13068-019-1549-x. eCollection 2019. Biotechnol Biofuels. 2019. PMID: 31497068 Free PMC article.
-
Biotechnological applications of mobile group II introns and their reverse transcriptases: gene targeting, RNA-seq, and non-coding RNA analysis.Mob DNA. 2014 Jan 13;5(1):2. doi: 10.1186/1759-8753-5-2. Mob DNA. 2014. PMID: 24410776 Free PMC article.
-
Mechanisms involved in xyloglucan catabolism by the cellulosome-producing bacterium Ruminiclostridium cellulolyticum.Sci Rep. 2016 Mar 7;6:22770. doi: 10.1038/srep22770. Sci Rep. 2016. PMID: 26946939 Free PMC article.
-
Handling Several Sugars at a Time: a Case Study of Xyloglucan Utilization by Ruminiclostridium cellulolyticum.mBio. 2021 Dec 21;12(6):e0220621. doi: 10.1128/mBio.02206-21. Epub 2021 Nov 9. mBio. 2021. PMID: 34749527 Free PMC article.
References
-
- Himmel ME, Ding SY, Johnson DK, Adney WS, Nimlos MR, et al. (2007) Biomass recalcitrance: engineering plants and enzymes for biofuels production. Science 315: 804–807. - PubMed
-
- Jordan DB, Bowman MJ, Braker JD, Dien BS, Hector RE, et al. (2012) Plant cell walls to ethanol. Biochem J 442: 241–252. - PubMed
-
- Petitdemange E, Caillet F, Giallo J, Gaudin C (1984) Clostridium cellulolyticum sp. nov., a cellulolytic mesophilic species from decayed grass. Int J Syst Bacteriol 34: 155–159.
-
- Li Y, Tschaplinski TJ, Engle NL, Hamilton CY, Rodriguez M Jr, et al. (2012) Combined inactivation of the Clostridium cellulolyticum lactate and malate dehydrogenase genes substantially increases ethanol yield from cellulose and switchgrass fermentations. Biotechnology for biofuels 5: 2. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

