Beyond secondary structure: primary-sequence determinants license pri-miRNA hairpins for processing
- PMID: 23415231
- PMCID: PMC3707628
- DOI: 10.1016/j.cell.2013.01.031
Beyond secondary structure: primary-sequence determinants license pri-miRNA hairpins for processing
Abstract
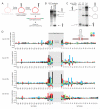
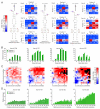
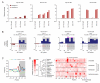
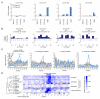
To use microRNAs to downregulate mRNA targets, cells must first process these ~22 nt RNAs from primary transcripts (pri-miRNAs). These transcripts form RNA hairpins important for processing, but additional determinants must distinguish pri-miRNAs from the many other hairpin-containing transcripts expressed in each cell. Illustrating the complexity of this recognition, we show that most Caenorhabditis elegans pri-miRNAs lack determinants required for processing in human cells. To find these determinants, we generated many variants of four human pri-miRNAs, sequenced millions that retained function, and compared them with the starting variants. Our results confirmed the importance of pairing in the stem and revealed three primary-sequence determinants, including an SRp20-binding motif (CNNC) found downstream of most pri-miRNA hairpins in bilaterian animals, but not in nematodes. Adding this and other determinants to C. elegans pri-miRNAs imparted efficient processing in human cells, thereby confirming the importance of primary-sequence determinants for distinguishing pri-miRNAs from other hairpin-containing transcripts.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures



Similar articles
-
Novel determinants of mammalian primary microRNA processing revealed by systematic evaluation of hairpin-containing transcripts and human genetic variation.Genome Res. 2017 Mar;27(3):374-384. doi: 10.1101/gr.208900.116. Epub 2017 Jan 13. Genome Res. 2017. PMID: 28087842 Free PMC article.
-
SRSF3 recruits DROSHA to the basal junction of primary microRNAs.RNA. 2018 Jul;24(7):892-898. doi: 10.1261/rna.065862.118. Epub 2018 Apr 3. RNA. 2018. PMID: 29615481 Free PMC article.
-
Splicing remodels the let-7 primary microRNA to facilitate Drosha processing in Caenorhabditis elegans.RNA. 2015 Aug;21(8):1396-403. doi: 10.1261/rna.052118.115. Epub 2015 Jun 16. RNA. 2015. PMID: 26081559 Free PMC article.
-
Orientation of Human Microprocessor on Primary MicroRNAs.Biochemistry. 2019 Jan 29;58(4):189-198. doi: 10.1021/acs.biochem.8b00944. Epub 2018 Dec 10. Biochemistry. 2019. PMID: 30481000 Review.
-
New insights into pri-miRNA processing and accumulation in plants.Wiley Interdiscip Rev RNA. 2015 Sep-Oct;6(5):533-45. doi: 10.1002/wrna.1292. Epub 2015 Jun 29. Wiley Interdiscip Rev RNA. 2015. PMID: 26119101 Review.
Cited by
-
N6-methyladenosine marks primary microRNAs for processing.Nature. 2015 Mar 26;519(7544):482-5. doi: 10.1038/nature14281. Epub 2015 Mar 18. Nature. 2015. PMID: 25799998 Free PMC article.
-
Next-generation libraries for robust RNA interference-based genome-wide screens.Proc Natl Acad Sci U S A. 2015 Jun 30;112(26):E3384-91. doi: 10.1073/pnas.1508821112. Epub 2015 Jun 15. Proc Natl Acad Sci U S A. 2015. PMID: 26080438 Free PMC article.
-
The cellular and KSHV A-to-I RNA editome in primary effusion lymphoma and its role in the viral lifecycle.Nat Commun. 2023 Mar 13;14(1):1367. doi: 10.1038/s41467-023-37105-8. Nat Commun. 2023. PMID: 36914661 Free PMC article.
-
pH dependence of C•A, G•A and A•A mismatches in the stem of precursor microRNA-31.Biophys Chem. 2022 Apr;283:106763. doi: 10.1016/j.bpc.2022.106763. Epub 2022 Jan 22. Biophys Chem. 2022. PMID: 35114594 Free PMC article.
-
MirGeneDB 2.0: the metazoan microRNA complement.Nucleic Acids Res. 2020 Jan 8;48(D1):D132-D141. doi: 10.1093/nar/gkz885. Nucleic Acids Res. 2020. PMID: 31598695 Free PMC article.
References
-
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
-
- Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, Barzilai A, Einat P, Einav U, Meiri E, et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005;37:766–770. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

