Concise review: new paradigms for Down syndrome research using induced pluripotent stem cells: tackling complex human genetic disease
- PMID: 23413375
- PMCID: PMC3659762
- DOI: 10.5966/sctm.2012-0117
Concise review: new paradigms for Down syndrome research using induced pluripotent stem cells: tackling complex human genetic disease
Abstract
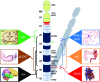
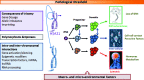
Down syndrome (DS) is a complex developmental disorder with diverse pathologies that affect multiple tissues and organ systems. Clear mechanistic description of how trisomy of chromosome 21 gives rise to most DS pathologies is currently lacking and is limited to a few examples of dosage-sensitive trisomic genes with large phenotypic effects. The recent advent of cellular reprogramming technology offers a promising way forward, by allowing derivation of patient-derived human cell types in vitro. We present general strategies that integrate genomics technologies and induced pluripotent stem cells to identify molecular networks driving different aspects of DS pathogenesis and describe experimental approaches to validate the causal requirement of candidate network defects for particular cellular phenotypes. This overall approach should be applicable to many poorly understood complex human genetic diseases, whose pathogenic mechanisms might involve the combined effects of many genes.
Figures
Similar articles
-
Integration-free induced pluripotent stem cells model genetic and neural developmental features of down syndrome etiology.Stem Cells. 2013 Mar;31(3):467-78. doi: 10.1002/stem.1297. Stem Cells. 2013. PMID: 23225669
-
Generation of two induced pluripotent stem cell lines from patients with Down syndrome.Stem Cell Res. 2023 Oct;72:103204. doi: 10.1016/j.scr.2023.103204. Epub 2023 Sep 14. Stem Cell Res. 2023. PMID: 37734318 Free PMC article.
-
Mental retardation in Down syndrome: from gene dosage imbalance to molecular and cellular mechanisms.Neurosci Res. 2007 Dec;59(4):349-69. doi: 10.1016/j.neures.2007.08.007. Epub 2007 Aug 15. Neurosci Res. 2007. PMID: 17897742 Review.
-
Generation of Integration-Free Induced Pluripotent Stem Cells from Urine-Derived Cells Isolated from Individuals with Down Syndrome.Stem Cells Transl Med. 2017 Jun;6(6):1465-1476. doi: 10.1002/sctm.16-0128. Epub 2017 Mar 28. Stem Cells Transl Med. 2017. PMID: 28371411 Free PMC article.
-
Developmental instability of the cerebellum and its relevance to Down syndrome.J Neural Transm Suppl. 2001;(61):11-34. doi: 10.1007/978-3-7091-6262-0_2. J Neural Transm Suppl. 2001. PMID: 11771737 Review.
Cited by
-
Molecular Characterization of Down Syndrome Embryonic Stem Cells Reveals a Role for RUNX1 in Neural Differentiation.Stem Cell Reports. 2016 Oct 11;7(4):777-786. doi: 10.1016/j.stemcr.2016.08.003. Epub 2016 Sep 8. Stem Cell Reports. 2016. PMID: 27618722 Free PMC article.
-
Will brain cells derived from induced pluripotent stem cells or directly converted from somatic cells (iNs) be useful for schizophrenia research?Schizophr Bull. 2013 Sep;39(5):948-54. doi: 10.1093/schbul/sbt103. Epub 2013 Jul 24. Schizophr Bull. 2013. PMID: 23884351 Free PMC article. Review.
-
Trisomy 21 is Associated with Caspase-2 Upregulation in Cytotrophoblasts at the Maternal-Fetal Interface.Reprod Sci. 2020 Jan;27(1):100-109. doi: 10.1007/s43032-019-00002-x. Epub 2020 Jan 1. Reprod Sci. 2020. PMID: 32046398 Free PMC article.
-
iPS Cells-The Triumphs and Tribulations.Dent J (Basel). 2016 Jun 6;4(2):19. doi: 10.3390/dj4020019. Dent J (Basel). 2016. PMID: 29563461 Free PMC article. Review.
-
"Cutting the Mustard" with Induced Pluripotent Stem Cells: An Overview and Applications in Healthcare Paradigm.Stem Cell Rev Rep. 2022 Dec;18(8):2757-2780. doi: 10.1007/s12015-022-10390-4. Epub 2022 Jul 6. Stem Cell Rev Rep. 2022. PMID: 35793037 Review.
References
-
- Parker SE, Mai CT, Canfield MA, et al. Updated National Birth Prevalence estimates for selected birth defects in the United States, 2004–2006. Birth Defects Res A Clin Mol Teratol. 2010;88:1008–1016. - PubMed
-
- Hassold T, Hall H, Hunt P. The origin of human aneuploidy: Where we have been, where we are going. Hum Mol Genet. 2007;16(Spec No. 2):R203–R208. - PubMed
-
- Petersen MB, Mikkelsen M. Nondisjunction in trisomy 21: Origin and mechanisms. Cytogenet Cell Genet. 2000;91:199–203. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

