Inferring admixture histories of human populations using linkage disequilibrium
- PMID: 23410830
- PMCID: PMC3606100
- DOI: 10.1534/genetics.112.147330
Inferring admixture histories of human populations using linkage disequilibrium
Abstract
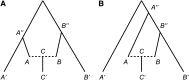
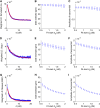
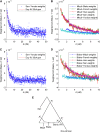
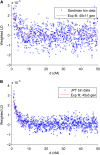
Long-range migrations and the resulting admixtures between populations have been important forces shaping human genetic diversity. Most existing methods for detecting and reconstructing historical admixture events are based on allele frequency divergences or patterns of ancestry segments in chromosomes of admixed individuals. An emerging new approach harnesses the exponential decay of admixture-induced linkage disequilibrium (LD) as a function of genetic distance. Here, we comprehensively develop LD-based inference into a versatile tool for investigating admixture. We present a new weighted LD statistic that can be used to infer mixture proportions as well as dates with fewer constraints on reference populations than previous methods. We define an LD-based three-population test for admixture and identify scenarios in which it can detect admixture events that previous formal tests cannot. We further show that we can uncover phylogenetic relationships among populations by comparing weighted LD curves obtained using a suite of references. Finally, we describe several improvements to the computation and fitting of weighted LD curves that greatly increase the robustness and speed of the calculations. We implement all of these advances in a software package, ALDER, which we validate in simulations and apply to test for admixture among all populations from the Human Genome Diversity Project (HGDP), highlighting insights into the admixture history of Central African Pygmies, Sardinians, and Japanese.
Figures
Similar articles
-
Inference of multiple-wave population admixture by modeling decay of linkage disequilibrium with polynomial functions.Heredity (Edinb). 2017 May;118(5):503-510. doi: 10.1038/hdy.2017.5. Epub 2017 Feb 15. Heredity (Edinb). 2017. PMID: 28198814 Free PMC article.
-
Estimating the timing of multiple admixture events using 3-locus linkage disequilibrium.PLoS Genet. 2022 Jul 15;18(7):e1010281. doi: 10.1371/journal.pgen.1010281. eCollection 2022 Jul. PLoS Genet. 2022. PMID: 35839249 Free PMC article.
-
Complex genetic admixture histories reconstructed with Approximate Bayesian Computation.Mol Ecol Resour. 2021 May;21(4):1098-1117. doi: 10.1111/1755-0998.13325. Epub 2021 Feb 26. Mol Ecol Resour. 2021. PMID: 33452723 Free PMC article.
-
Mapping asthma-associated variants in admixed populations.Front Genet. 2015 Sep 29;6:292. doi: 10.3389/fgene.2015.00292. eCollection 2015. Front Genet. 2015. PMID: 26483834 Free PMC article. Review.
-
Admixture mapping and the role of population structure for localizing disease genes.Adv Genet. 2008;60:547-69. doi: 10.1016/S0065-2660(07)00419-1. Adv Genet. 2008. PMID: 18358332 Review.
Cited by
-
Signatures of Convergent Evolution and Natural Selection at the Alcohol Dehydrogenase Gene Region are Correlated with Agriculture in Ethnically Diverse Africans.Mol Biol Evol. 2022 Oct 7;39(10):msac183. doi: 10.1093/molbev/msac183. Mol Biol Evol. 2022. PMID: 36026493 Free PMC article.
-
Ancient Rapanui genomes reveal resilience and pre-European contact with the Americas.Nature. 2024 Sep;633(8029):389-397. doi: 10.1038/s41586-024-07881-4. Epub 2024 Sep 11. Nature. 2024. PMID: 39261618 Free PMC article.
-
A Genome-Wide Search for Greek and Jewish Admixture in the Kashmiri Population.PLoS One. 2016 Aug 4;11(8):e0160614. doi: 10.1371/journal.pone.0160614. eCollection 2016. PLoS One. 2016. PMID: 27490348 Free PMC article.
-
ALDsuite: Dense marker MALD using principal components of ancestral linkage disequilibrium.BMC Genet. 2015 Mar 7;16:23. doi: 10.1186/s12863-015-0179-y. BMC Genet. 2015. PMID: 25886794 Free PMC article.
-
Genetic evidence for a single founding population of the Lakshadweep Islands.Mol Genet Genomics. 2024 Feb 19;299(1):8. doi: 10.1007/s00438-024-02110-z. Mol Genet Genomics. 2024. PMID: 38374307
References
-
- Bramanti B., Thomas M., Haak W., Unterlaender M., Jores P., et al. , 2009. Genetic discontinuity between local hunter–gatherers and Central Europe’s first farmers. Science 326(5949): 137–140. - PubMed
-
- Davies R., 1977. Hypothesis testing when a nuisance parameter is present only under the alternative. Biometrika 64(2): 247–254. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

