Antagonism versus cooperativity with TALE cofactors at the base of the functional diversification of Hox protein function
- PMID: 23408901
- PMCID: PMC3567137
- DOI: 10.1371/journal.pgen.1003252
Antagonism versus cooperativity with TALE cofactors at the base of the functional diversification of Hox protein function
Abstract
Extradenticle (Exd) and Homothorax (Hth) function as positive transcriptional cofactors of Hox proteins, helping them to bind specifically their direct targets. The posterior Hox protein Abdominal-B (Abd-B) does not require Exd/Hth to bind DNA; and, during embryogenesis, Abd-B represses hth and exd transcription. Here we show that this repression is necessary for Abd-B function, as maintained Exd/Hth expression results in transformations similar to those observed in loss-of-function Abd-B mutants. We characterize the cis regulatory module directly regulated by Abd-B in the empty spiracles gene and show that the Exd/Hth complex interferes with Abd-B binding to this enhancer. Our results suggest that this novel Exd/Hth function does not require the complex to bind DNA and may be mediated by direct Exd/Hth binding to the Abd-B homeodomain. Thus, in some instances, the main positive cofactor complex for anterior Hox proteins can act as a negative factor for the posterior Hox protein Abd-B. This antagonistic interaction uncovers an alternative way in which MEIS and PBC cofactors can modulate Abd-B like posterior Hox genes during development.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


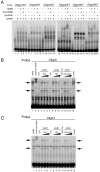
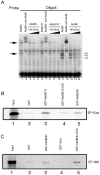
Similar articles
-
The cis-regulatory logic underlying abdominal Hox-mediated repression versus activation of regulatory elements in Drosophila.Dev Biol. 2019 Jan 15;445(2):226-236. doi: 10.1016/j.ydbio.2018.11.006. Epub 2018 Nov 20. Dev Biol. 2019. PMID: 30468713 Free PMC article.
-
Hox repression of a target gene: extradenticle-independent, additive action through multiple monomer binding sites.Development. 2002 Jul;129(13):3115-26. doi: 10.1242/dev.129.13.3115. Development. 2002. PMID: 12070087
-
Ambivalent partnership of the Drosophila posterior class Hox protein Abdominal-B with Extradenticle and Homothorax.PLoS Genet. 2025 Jan 13;21(1):e1011355. doi: 10.1371/journal.pgen.1011355. eCollection 2025 Jan. PLoS Genet. 2025. PMID: 39804927 Free PMC article.
-
Hox cofactors in vertebrate development.Dev Biol. 2006 Mar 15;291(2):193-206. doi: 10.1016/j.ydbio.2005.10.032. Epub 2006 Mar 3. Dev Biol. 2006. PMID: 16515781 Review.
-
Cellular analysis of newly identified Hox downstream genes in Drosophila.Eur J Cell Biol. 2010 Feb-Mar;89(2-3):273-8. doi: 10.1016/j.ejcb.2009.11.012. Epub 2009 Dec 16. Eur J Cell Biol. 2010. PMID: 20018403 Review.
Cited by
-
Chromatin accessibility plays a key role in selective targeting of Hox proteins.Genome Biol. 2019 Jun 3;20(1):115. doi: 10.1186/s13059-019-1721-4. Genome Biol. 2019. PMID: 31159833 Free PMC article.
-
Hox Proteins Act as Transcriptional Guarantors to Ensure Terminal Differentiation.Cell Rep. 2015 Nov 17;13(7):1343-1352. doi: 10.1016/j.celrep.2015.10.044. Epub 2015 Nov 4. Cell Rep. 2015. PMID: 26547238 Free PMC article.
-
Role of HOXA9 in leukemia: dysregulation, cofactors and essential targets.Oncogene. 2016 Mar 3;35(9):1090-8. doi: 10.1038/onc.2015.174. Epub 2015 Jun 1. Oncogene. 2016. PMID: 26028034 Free PMC article. Review.
-
Abdominal-B and caudal inhibit the formation of specific neuroblasts in the Drosophila tail region.Development. 2013 Sep;140(17):3552-64. doi: 10.1242/dev.096099. Epub 2013 Jul 31. Development. 2013. PMID: 23903193 Free PMC article.
-
Transcriptional Regulation and Implications for Controlling Hox Gene Expression.J Dev Biol. 2022 Jan 10;10(1):4. doi: 10.3390/jdb10010004. J Dev Biol. 2022. PMID: 35076545 Free PMC article. Review.
References
-
- Pearson JC, Lemons D, McGinnis W (2005) Modulating Hox gene functions during animal body patterning. Nat Rev Genet 6: 893–904. - PubMed
-
- Hueber SD, Weiller GF, Djordjevic MA, Frickey T (2010) Improving Hox protein classification across the major model organisms. PLoS ONE 5: e10820 doi:10.1371/journal.pone.0010820. - DOI - PMC - PubMed
-
- Merabet S, Hudry B, Saadaoui M, Graba Y (2009) Classification of sequence signatures: a guide to Hox protein function. Bioessays 31: 500–511. - PubMed
-
- Hombría JC-G, Rivas ML, Sotillos S (2009) Genetic control of morphogenesis- Hox induced organogenesis of the posterior spiracles. Int J Dev Biol 53: 1349–1358. - PubMed
-
- Hu N, Castelli-Gair J (1999) Study of the posterior spiracles of Drosophila as a model to understand the genetic and cellular mechanisms controlling morphogenesis. Developmental Biology 214: 197–210. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

