Quantitative analysis of TALE-DNA interactions suggests polarity effects
- PMID: 23408851
- PMCID: PMC3627578
- DOI: 10.1093/nar/gkt085
Quantitative analysis of TALE-DNA interactions suggests polarity effects
Abstract
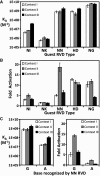
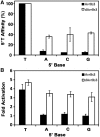
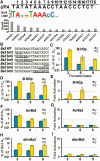
Transcription activator-like effectors (TALEs) have revolutionized the field of genome engineering. We present here a systematic assessment of TALE DNA recognition, using quantitative electrophoretic mobility shift assays and reporter gene activation assays. Within TALE proteins, tandem 34-amino acid repeats recognize one base pair each and direct sequence-specific DNA binding through repeat variable di-residues (RVDs). We found that RVD choice can affect affinity by four orders of magnitude, with the relative RVD contribution in the order NG > HD ≈ NN >> NI > NK. The NN repeat preferred the base G over A, whereas the NK repeat bound G with 10(3)-fold lower affinity. We compared AvrBs3, a naturally occurring TALE that recognizes its target using some atypical RVD-base combinations, with a designed TALE that precisely matches 'standard' RVDs with the target bases. This comparison revealed unexpected differences in sensitivity to substitutions of the invariant 5'-T. Another surprising observation was that base mismatches at the 5' end of the target site had more disruptive effects on affinity than those at the 3' end, particularly in designed TALEs. These results provide evidence that TALE-DNA recognition exhibits a hitherto un-described polarity effect, in which the N-terminal repeats contribute more to affinity than C-terminal ones.
Figures

Similar articles
-
Programmable DNA-binding proteins from Burkholderia provide a fresh perspective on the TALE-like repeat domain.Nucleic Acids Res. 2014 Jun;42(11):7436-49. doi: 10.1093/nar/gku329. Epub 2014 May 3. Nucleic Acids Res. 2014. PMID: 24792163 Free PMC article.
-
Targeting G with TAL effectors: a comparison of activities of TALENs constructed with NN and NK repeat variable di-residues.PLoS One. 2012;7(9):e45383. doi: 10.1371/journal.pone.0045383. Epub 2012 Sep 24. PLoS One. 2012. PMID: 23028976 Free PMC article.
-
Repeat 1 of TAL effectors affects target specificity for the base at position zero.Nucleic Acids Res. 2014 Jun;42(11):7160-9. doi: 10.1093/nar/gku341. Epub 2014 May 3. Nucleic Acids Res. 2014. PMID: 24792160 Free PMC article.
-
Conformational elasticity can facilitate TALE-DNA recognition.Adv Protein Chem Struct Biol. 2014;94:347-64. doi: 10.1016/B978-0-12-800168-4.00009-3. Adv Protein Chem Struct Biol. 2014. PMID: 24629191 Free PMC article. Review.
-
TALE: a tale of genome editing.Prog Biophys Mol Biol. 2014 Jan;114(1):25-32. doi: 10.1016/j.pbiomolbio.2013.11.006. Epub 2013 Nov 27. Prog Biophys Mol Biol. 2014. PMID: 24291598 Review.
Cited by
-
Biotechnological strategies and tools for Plum pox virus resistance: trans-, intra-, cis-genesis, and beyond.Front Plant Sci. 2015 Jun 8;6:379. doi: 10.3389/fpls.2015.00379. eCollection 2015. Front Plant Sci. 2015. PMID: 26106397 Free PMC article. Review.
-
TALEN-mediated genome engineering to generate targeted mice.Chromosome Res. 2015 Feb;23(1):43-55. doi: 10.1007/s10577-014-9457-1. Chromosome Res. 2015. PMID: 25596827 Review.
-
PrediTALE: A novel model learned from quantitative data allows for new perspectives on TALE targeting.PLoS Comput Biol. 2019 Jul 11;15(7):e1007206. doi: 10.1371/journal.pcbi.1007206. eCollection 2019 Jul. PLoS Comput Biol. 2019. PMID: 31295249 Free PMC article.
-
TAL effector driven induction of a SWEET gene confers susceptibility to bacterial blight of cotton.Nat Commun. 2017 May 24;8:15588. doi: 10.1038/ncomms15588. Nat Commun. 2017. PMID: 28537271 Free PMC article.
-
Context influences on TALE-DNA binding revealed by quantitative profiling.Nat Commun. 2015 Jun 11;6:7440. doi: 10.1038/ncomms8440. Nat Commun. 2015. PMID: 26067805 Free PMC article.
References
-
- Boch J, Bonas U. Xanthomonas AvrBs3 Family-Type III Effectors: Discovery and Function. Annu. Rev. Phytopathol. 2010;48:419–436. - PubMed
-
- Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509–1512. - PubMed
-
- Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326:1501. - PubMed

