Histone chaperones Nap1 and Vps75 regulate histone acetylation during transcription elongation
- PMID: 23401858
- PMCID: PMC3624251
- DOI: 10.1128/MCB.01121-12
Histone chaperones Nap1 and Vps75 regulate histone acetylation during transcription elongation
Abstract
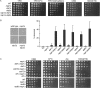
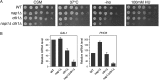
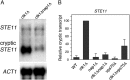
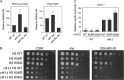
Histone chaperones function in chromatin assembly and disassembly, suggesting they have important regulatory roles in transcription elongation. The Saccharomyces cerevisiae proteins Nap1 and Vps75 are structurally related, evolutionarily conserved histone chaperones. We showed that Nap1 genetically interacts with several transcription elongation factors and that both Nap1 and Vps75 interact with the RNA polymerase II kinase, CTK1. Loss of NAP1 or VPS75 suppressed cryptic transcription within the open reading frame (ORF) observed when strains are deleted for the kinase CTK1. Loss of the histone acetyltransferase Rtt109 also suppressed ctk1-dependent cryptic transcription. Vps75 regulates Rtt109 function, suggesting that they function together in this process. Histone H3 K9 was found to be the important lysine that is acetylated by Rtt109 during ctk1-dependent cryptic transcription. We showed that both Vps75 and Nap1 regulate the relative level of H3 K9 acetylation in the STE11 ORF. This supports a model in which Nap1, like Vps75, directly regulates Rtt109 activity or regulates the assembly of acetylated chromatin. Although Nap1 and Vps75 share many similarities, due to their distinct interactions with SET2, Nap1 and Vps75 may also play separate roles during transcription elongation. This work sheds further light on the importance of histone chaperones as general regulators of transcription elongation.
Figures



Similar articles
-
Interaction with the histone chaperone Vps75 promotes nuclear localization and HAT activity of Rtt109 in vivo.Traffic. 2011 Jul;12(7):826-39. doi: 10.1111/j.1600-0854.2011.01202.x. Epub 2011 May 5. Traffic. 2011. PMID: 21463458 Free PMC article.
-
Molecular functions of the histone acetyltransferase chaperone complex Rtt109-Vps75.Nat Struct Mol Biol. 2008 Sep;15(9):948-56. doi: 10.1038/nsmb.1459. Nat Struct Mol Biol. 2008. PMID: 19172748 Free PMC article.
-
Structure of the Rtt109-AcCoA/Vps75 complex and implications for chaperone-mediated histone acetylation.Structure. 2011 Feb 9;19(2):221-31. doi: 10.1016/j.str.2010.12.012. Epub 2011 Jan 20. Structure. 2011. PMID: 21256037 Free PMC article.
-
Understanding histone acetyltransferase Rtt109 structure and function: how many chaperones does it take?Curr Opin Struct Biol. 2011 Dec;21(6):728-34. doi: 10.1016/j.sbi.2011.09.005. Epub 2011 Oct 23. Curr Opin Struct Biol. 2011. PMID: 22023828 Free PMC article. Review.
-
Histone-modifying enzymes, histone modifications and histone chaperones in nucleosome assembly: Lessons learned from Rtt109 histone acetyltransferases.Crit Rev Biochem Mol Biol. 2015 Jan-Feb;50(1):31-53. doi: 10.3109/10409238.2014.978975. Epub 2014 Nov 3. Crit Rev Biochem Mol Biol. 2015. PMID: 25365782 Free PMC article. Review.
Cited by
-
Histone chaperone networks shaping chromatin function.Nat Rev Mol Cell Biol. 2017 Mar;18(3):141-158. doi: 10.1038/nrm.2016.159. Epub 2017 Jan 5. Nat Rev Mol Cell Biol. 2017. PMID: 28053344 Free PMC article. Review.
-
The histone chaperones Vps75 and Nap1 form ring-like, tetrameric structures in solution.Nucleic Acids Res. 2014 May;42(9):6038-51. doi: 10.1093/nar/gku232. Epub 2014 Mar 31. Nucleic Acids Res. 2014. PMID: 24688059 Free PMC article.
-
The histone chaperone Vps75 forms multiple oligomeric assemblies capable of mediating exchange between histone H3-H4 tetramers and Asf1-H3-H4 complexes.Nucleic Acids Res. 2016 Jul 27;44(13):6157-72. doi: 10.1093/nar/gkw209. Epub 2016 Apr 1. Nucleic Acids Res. 2016. PMID: 27036862 Free PMC article.
-
The histone chaperone NAP1L3 is required for haematopoietic stem cell maintenance and differentiation.Sci Rep. 2018 Jul 25;8(1):11202. doi: 10.1038/s41598-018-29518-z. Sci Rep. 2018. PMID: 30046127 Free PMC article.
-
The histone chaperone FACT: a guardian of chromatin structure integrity.Transcription. 2022 Feb-Jun;13(1-3):16-38. doi: 10.1080/21541264.2022.2069995. Epub 2022 Apr 29. Transcription. 2022. PMID: 35485711 Free PMC article. Review.
References
-
- Zlatanova J, Seebart C, Tomschik M. 2007. Nap1: taking a closer look at a juggler protein of extraordinary skills. FASEB J. 21:1294–1310 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
