Time-resolved fluorescence imaging reveals differential interactions of N-glycan processing enzymes across the Golgi stack in planta
- PMID: 23400704
- PMCID: PMC3613452
- DOI: 10.1104/pp.112.210757
Time-resolved fluorescence imaging reveals differential interactions of N-glycan processing enzymes across the Golgi stack in planta
Abstract
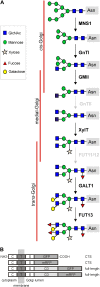
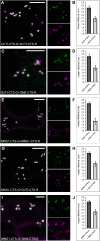
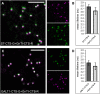
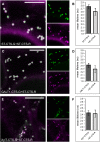
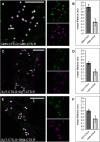
N-Glycan processing is one of the most important cellular protein modifications in plants and as such is essential for plant development and defense mechanisms. The accuracy of Golgi-located processing steps is governed by the strict intra-Golgi localization of sequentially acting glycosidases and glycosyltransferases. Their differential distribution goes hand in hand with the compartmentalization of the Golgi stack into cis-, medial-, and trans-cisternae, which separate early from late processing steps. The mechanisms that direct differential enzyme concentration are still unknown, but the formation of multienzyme complexes is considered a feasible Golgi protein localization strategy. In this study, we used two-photon excitation-Förster resonance energy transfer-fluorescence lifetime imaging microscopy to determine the interaction of N-glycan processing enzymes with differential intra-Golgi locations. Following the coexpression of fluorescent protein-tagged amino-terminal Golgi-targeting sequences (cytoplasmic-transmembrane-stem [CTS] region) of enzyme pairs in leaves of tobacco (Nicotiana spp.), we observed that all tested cis- and medial-Golgi enzymes, namely Arabidopsis (Arabidopsis thaliana) Golgi α-mannosidase I, Nicotiana tabacum β1,2-N-acetylglucosaminyltransferase I, Arabidopsis Golgi α-mannosidase II (GMII), and Arabidopsis β1,2-xylosyltransferase, form homodimers and heterodimers, whereas among the late-acting enzymes Arabidopsis β1,3-galactosyltransferase1 (GALT1), Arabidopsis α1,4-fucosyltransferase, and Rattus norvegicus α2,6-sialyltransferase (a nonplant Golgi marker), only GALT1 and medial-Golgi GMII were found to form a heterodimer. Furthermore, the efficiency of energy transfer indicating the formation of interactions decreased considerably in a cis-to-trans fashion. The comparative fluorescence lifetime imaging of several full-length cis- and medial-Golgi enzymes and their respective catalytic domain-deleted CTS clones further suggested that the formation of protein-protein interactions can occur through their amino-terminal CTS region.
Figures
Similar articles
-
Plant N-glycan processing enzymes employ different targeting mechanisms for their spatial arrangement along the secretory pathway.Plant Cell. 2006 Nov;18(11):3182-200. doi: 10.1105/tpc.105.036400. Epub 2006 Nov 30. Plant Cell. 2006. PMID: 17138701 Free PMC article.
-
The Golgi Localization of GnTI Requires a Polar Amino Acid Residue within Its Transmembrane Domain.Plant Physiol. 2019 Jun;180(2):859-873. doi: 10.1104/pp.19.00310. Epub 2019 Apr 10. Plant Physiol. 2019. PMID: 30971450 Free PMC article.
-
Alpha-mannosidases involved in N-glycan processing show cell specificity and distinct subcompartmentalization within the Golgi apparatus of cells in the testis and epididymis.Eur J Cell Biol. 1999 Jul;78(7):441-52. doi: 10.1016/s0171-9335(99)80071-5. Eur J Cell Biol. 1999. PMID: 10472797
-
Sub-compartmental organization of Golgi-resident N-glycan processing enzymes in plants.Mol Plant. 2011 Mar;4(2):220-8. doi: 10.1093/mp/ssq082. Epub 2011 Feb 9. Mol Plant. 2011. PMID: 21307368 Free PMC article. Review.
-
Golgi localization of glycosyltransferases: more questions than answers.Glycobiology. 1997 Feb;7(1):1-13. doi: 10.1093/glycob/7.1.1-b. Glycobiology. 1997. PMID: 9061359 Free PMC article. Review.
Cited by
-
Identification of a novel Golgi-localized putative glycosyltransferase protein in Arabidopsis thaliana.Plant Biotechnol (Tokyo). 2024 Mar 25;41(1):35-44. doi: 10.5511/plantbiotechnology.23.1214a. Plant Biotechnol (Tokyo). 2024. PMID: 39464868 Free PMC article.
-
UDP-galactose (SLC35A2) and UDP-N-acetylglucosamine (SLC35A3) Transporters Form Glycosylation-related Complexes with Mannoside Acetylglucosaminyltransferases (Mgats).J Biol Chem. 2015 Jun 19;290(25):15475-15486. doi: 10.1074/jbc.M115.636670. Epub 2015 May 5. J Biol Chem. 2015. PMID: 25944901 Free PMC article.
-
Processing of the Terminal Alpha-1,2-Linked Mannose Residues From Oligomannosidic N-Glycans Is Critical for Proper Root Growth.Front Plant Sci. 2018 Dec 6;9:1807. doi: 10.3389/fpls.2018.01807. eCollection 2018. Front Plant Sci. 2018. PMID: 30574158 Free PMC article.
-
Galactosyltransferases from Arabidopsis thaliana in the biosynthesis of type II arabinogalactan: molecular interaction enhances enzyme activity.BMC Plant Biol. 2014 Apr 3;14:90. doi: 10.1186/1471-2229-14-90. BMC Plant Biol. 2014. PMID: 24693939 Free PMC article.
-
Endomembrane and Golgi traffic in plant cells.Methods Cell Biol. 2013;118:69-83. doi: 10.1016/B978-0-12-417164-0.00005-7. Methods Cell Biol. 2013. PMID: 24295301 Free PMC article.
References
-
- Adjobo-Hermans MJ, Goedhart J, Gadella TW., Jr (2006) Plant G protein heterotrimers require dual lipidation motifs of Galpha and Ggamma and do not dissociate upon activation. J Cell Sci 119: 5087–5097 - PubMed
-
- Aker J, Hesselink R, Engel R, Karlova R, Borst JW, Visser AJ, de Vries SC. (2007) In vivo hexamerization and characterization of the Arabidopsis AAA ATPase CDC48A complex using Forster resonance energy transfer-fluorescence lifetime imaging microscopy and fluorescence correlation spectroscopy. Plant Physiol 145: 339–350 - PMC - PubMed
-
- Atmodjo MA, Sakuragi Y, Zhu X, Burrell AJ, Mohanty SS, Atwood JA, III, Orlando R, Scheller HV, Mohnen D. (2011) Galacturonosyltransferase (GAUT)1 and GAUT7 are the core of a plant cell wall pectin biosynthetic homogalacturonan:galacturonosyltransferase complex. Proc Natl Acad Sci USA 108: 20225–20230 - PMC - PubMed
-
- Bastiaens PI, Squire A. (1999) Fluorescence lifetime imaging microscopy: spatial resolution of biochemical processes in the cell. Trends Cell Biol 9: 48–52 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

