A role for the RNA-binding protein MOS2 in microRNA maturation in Arabidopsis
- PMID: 23399598
- PMCID: PMC3641593
- DOI: 10.1038/cr.2013.23
A role for the RNA-binding protein MOS2 in microRNA maturation in Arabidopsis
Abstract
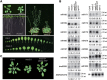
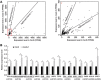
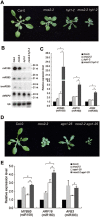
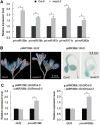
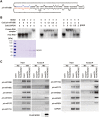
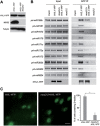
microRNAs (miRNAs) play important roles in the regulation of gene expression. In Arabidopsis, mature miRNAs are processed from primary miRNA transcripts (pri-miRNAs) by nuclear HYL1/SE/DCL1 complexes that form Dicing bodies (D-bodies). Here we report that an RNA-binding protein MOS2 binds to pri-miRNAs and is involved in efficient processing of pri-miRNAs. MOS2 does not interact with HYL1, SE, and DCL1 and is not localized in D-bodies. Interestingly, in the absence of MOS2, the recruitment of pri-miRNAs by HYL1 is greatly reduced and the localization of HYL1 in D-bodies is compromised. These data suggest that MOS2 promotes pri-miRNA processing through facilitating the recruitment of pri-miRNAs by the Dicing complexes.
Figures

Similar articles
-
Regulation of miRNA abundance by RNA binding protein TOUGH in Arabidopsis.Proc Natl Acad Sci U S A. 2012 Jul 31;109(31):12817-21. doi: 10.1073/pnas.1204915109. Epub 2012 Jul 16. Proc Natl Acad Sci U S A. 2012. PMID: 22802657 Free PMC article.
-
Homodimerization of HYL1 ensures the correct selection of cleavage sites in primary miRNA.Nucleic Acids Res. 2014 Oct 29;42(19):12224-36. doi: 10.1093/nar/gku907. Epub 2014 Oct 7. Nucleic Acids Res. 2014. PMID: 25294831 Free PMC article.
-
Identification of nuclear dicing bodies containing proteins for microRNA biogenesis in living Arabidopsis plants.Curr Biol. 2007 May 1;17(9):818-23. doi: 10.1016/j.cub.2007.04.005. Epub 2007 Apr 19. Curr Biol. 2007. PMID: 17442570 Free PMC article.
-
MicroRNA biogenesis and function in plants.FEBS Lett. 2005 Oct 31;579(26):5923-31. doi: 10.1016/j.febslet.2005.07.071. Epub 2005 Aug 9. FEBS Lett. 2005. PMID: 16144699 Free PMC article. Review.
-
HYL1's multiverse: A journey through miRNA biogenesis and beyond canonical and non-canonical functions of HYL1.Curr Opin Plant Biol. 2024 Aug;80:102546. doi: 10.1016/j.pbi.2024.102546. Epub 2024 May 7. Curr Opin Plant Biol. 2024. PMID: 38718678 Review.
Cited by
-
NPK macronutrients and microRNA homeostasis.Front Plant Sci. 2015 Jun 16;6:451. doi: 10.3389/fpls.2015.00451. eCollection 2015. Front Plant Sci. 2015. PMID: 26136763 Free PMC article. Review.
-
Complementation of HYPONASTIC LEAVES1 by double-strand RNA-binding domains of DICER-LIKE1 in nuclear dicing bodies.Plant Physiol. 2013 Sep;163(1):108-17. doi: 10.1104/pp.113.219071. Epub 2013 Jul 25. Plant Physiol. 2013. PMID: 23886622 Free PMC article.
-
GPKOW is essential for pre-mRNA splicing in vitro and suppresses splicing defect caused by dominant-negative DHX16 mutation in vivo.Biosci Rep. 2014 Dec 12;34(6):e00163. doi: 10.1042/BSR20140142. Biosci Rep. 2014. PMID: 25296192 Free PMC article.
-
The Arabidopsis MOS4-Associated Complex Promotes MicroRNA Biogenesis and Precursor Messenger RNA Splicing.Plant Cell. 2017 Oct;29(10):2626-2643. doi: 10.1105/tpc.17.00370. Epub 2017 Sep 25. Plant Cell. 2017. PMID: 28947490 Free PMC article.
-
Salt Stress Reveals a New Role for ARGONAUTE1 in miRNA Biogenesis at the Transcriptional and Posttranscriptional Levels.Plant Physiol. 2016 Sep;172(1):297-312. doi: 10.1104/pp.16.00830. Epub 2016 Jul 6. Plant Physiol. 2016. PMID: 27385819 Free PMC article.
References
-
- Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–385. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

