Ligand binding site identification by higher dimension molecular dynamics
- PMID: 23394112
- PMCID: PMC3706581
- DOI: 10.1021/ci300561b
Ligand binding site identification by higher dimension molecular dynamics
Abstract
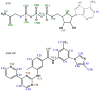
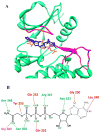
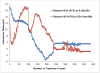
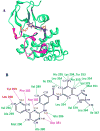
We propose a new molecular dynamics (MD) protocol to identify the binding site of a guest within a host. The method utilizes a four spatial (4D) dimension representation of the ligand allowing for rapid and efficient sampling within the receptor. We applied the method to two different model receptors characterized by diverse structural features of the binding site and different ligand binding affinities. The Abl kinase domain is comprised of a deep binding pocket and displays high affinity for the two chosen ligands examined here. The PDZ1 domain of PSD-95 has a shallow binding pocket that accommodates a peptide ligand involving far fewer interactions and a micromolar affinity. To ensure completely unbiased searching, the ligands were placed in the direct center of the protein receptors, away from the binding site, at the start of the 4D MD protocol. In both cases, the ligands were successfully docked into the binding site as identified in the published structures. The 4D MD protocol is able to overcome local energy barriers in locating the lowest energy binding pocket and will aid in the discovery of guest binding pockets in the absence of a priori knowledge of the site of interaction.
Figures
Similar articles
-
An in silico analysis of the binding modes and binding affinities of small molecule modulators of PDZ-peptide interactions.PLoS One. 2013 Aug 8;8(8):e71340. doi: 10.1371/journal.pone.0071340. eCollection 2013. PLoS One. 2013. PMID: 23951139 Free PMC article.
-
Ensemble-Based Analysis of the Dynamic Allostery in the PSD-95 PDZ3 Domain in Relation to the General Variability of PDZ Structures.Int J Mol Sci. 2020 Nov 6;21(21):8348. doi: 10.3390/ijms21218348. Int J Mol Sci. 2020. PMID: 33172212 Free PMC article.
-
Rigidified clicked dimeric ligands for studying the dynamics of the PDZ1-2 supramodule of PSD-95.Chembiochem. 2015 Jan 2;16(1):64-9. doi: 10.1002/cbic.201402547. Epub 2014 Nov 18. Chembiochem. 2015. PMID: 25407949
-
Simultaneous prediction of binding free energy and specificity for PDZ domain-peptide interactions.J Comput Aided Mol Des. 2013 Dec;27(12):1051-65. doi: 10.1007/s10822-013-9696-9. Epub 2013 Dec 5. J Comput Aided Mol Des. 2013. PMID: 24305904 Free PMC article.
-
Structural Analysis of Chemokine Receptor-Ligand Interactions.J Med Chem. 2017 Jun 22;60(12):4735-4779. doi: 10.1021/acs.jmedchem.6b01309. Epub 2017 Mar 10. J Med Chem. 2017. PMID: 28165741 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

