Multiple roles for Piwi in silencing Drosophila transposons
- PMID: 23392609
- PMCID: PMC3589557
- DOI: 10.1101/gad.209767.112
Multiple roles for Piwi in silencing Drosophila transposons
Abstract
Silencing of transposons in the Drosophila ovary relies on three Piwi family proteins--Piwi, Aubergine (Aub), and Ago3--acting in concert with their small RNA guides, the Piwi-interacting RNAs (piRNAs). Aub and Ago3 are found in the germ cell cytoplasm, where they function in the ping-pong cycle to consume transposon mRNAs. The nuclear Piwi protein is required for transposon silencing in both germ and somatic follicle cells, yet the precise mechanisms by which Piwi acts remain largely unclear. We investigated the role of Piwi by combining cell type-specific knockdowns with measurements of steady-state transposon mRNA levels, nascent RNA synthesis, chromatin state, and small RNA abundance. In somatic cells, Piwi loss led to concerted effects on nascent transcripts and transposon mRNAs, indicating that Piwi acts through transcriptional gene silencing (TGS). In germ cells, Piwi loss showed disproportionate impacts on steady-state RNA levels, indicating that it also exerts an effect on post-transcriptional gene silencing (PTGS). Piwi knockdown affected levels of germ cell piRNAs presumably bound to Aub and Ago3, perhaps explaining its post-transcriptional impacts. Overall, our results indicate that Piwi plays multiple roles in the piRNA pathway, in part enforcing transposon repression through effects on local chromatin states and transcription but also participating in germ cell piRNA biogenesis.
Figures
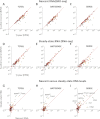

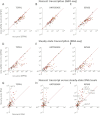
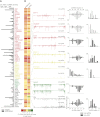
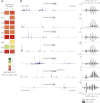
Comment in
-
Small RNA-directed silencing: the fly finds its inner fission yeast?Curr Biol. 2013 Apr 22;23(8):R318-20. doi: 10.1016/j.cub.2013.03.033. Curr Biol. 2013. PMID: 23618667
Similar articles
-
Heterotypic piRNA Ping-Pong requires qin, a protein with both E3 ligase and Tudor domains.Mol Cell. 2011 Nov 18;44(4):572-84. doi: 10.1016/j.molcel.2011.10.011. Mol Cell. 2011. PMID: 22099305 Free PMC article.
-
Slicing and Binding by Ago3 or Aub Trigger Piwi-Bound piRNA Production by Distinct Mechanisms.Mol Cell. 2015 Sep 3;59(5):819-30. doi: 10.1016/j.molcel.2015.08.007. Mol Cell. 2015. PMID: 26340424 Free PMC article.
-
[The interplay of transposon silencing genes in the Drosophila melanogaster germline].Mol Biol (Mosk). 2011 Jul-Aug;45(4):633-41. Mol Biol (Mosk). 2011. PMID: 21954595 Russian.
-
The piRNA pathway in Drosophila ovarian germ and somatic cells.Proc Jpn Acad Ser B Phys Biol Sci. 2020;96(1):32-42. doi: 10.2183/pjab.96.003. Proc Jpn Acad Ser B Phys Biol Sci. 2020. PMID: 31932527 Free PMC article. Review.
-
Untangling the web: the diverse functions of the PIWI/piRNA pathway.Mol Reprod Dev. 2013 Aug;80(8):632-64. doi: 10.1002/mrd.22195. Epub 2013 Jun 27. Mol Reprod Dev. 2013. PMID: 23712694 Free PMC article. Review.
Cited by
-
The emerging role of the piRNA/PIWI complex in respiratory tract diseases.Respir Res. 2023 Mar 13;24(1):76. doi: 10.1186/s12931-023-02367-9. Respir Res. 2023. PMID: 36915129 Free PMC article. Review.
-
PIWI proteins and PIWI-interacting RNAs in the soma.Nature. 2014 Jan 16;505(7483):353-359. doi: 10.1038/nature12987. Nature. 2014. PMID: 24429634 Free PMC article. Review.
-
Small RNAs in epigenetic inheritance: from mechanisms to trait transmission.FEBS Lett. 2021 Dec;595(24):2953-2977. doi: 10.1002/1873-3468.14210. Epub 2021 Oct 29. FEBS Lett. 2021. PMID: 34671979 Free PMC article. Review.
-
PID-1 is a novel factor that operates during 21U-RNA biogenesis in Caenorhabditis elegans.Genes Dev. 2014 Apr 1;28(7):683-8. doi: 10.1101/gad.238220.114. Genes Dev. 2014. PMID: 24696453 Free PMC article.
-
The Mi-2 nucleosome remodeler and the Rpd3 histone deacetylase are involved in piRNA-guided heterochromatin formation.Nat Commun. 2020 Jun 4;11(1):2818. doi: 10.1038/s41467-020-16635-5. Nat Commun. 2020. PMID: 32499524 Free PMC article.
References
-
- Aravin AA, Hannon GJ, Brennecke J 2007. The Piwi–piRNA pathway provides an adaptive defense in the transposon arms race. Science 318: 761–764 - PubMed
-
- Bregliano JC, Picard G, Bucheton A, Pelisson A, Lavige JM, L'Heritier P 1980. Hybrid dysgenesis in Drosophila melanogaster. Science 207: 606–611 - PubMed
-
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ 2007. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128: 1089–1103 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
