TGF-β inhibition enhances chemotherapy action against triple-negative breast cancer
- PMID: 23391723
- PMCID: PMC3582135
- DOI: 10.1172/JCI65416
TGF-β inhibition enhances chemotherapy action against triple-negative breast cancer
Abstract
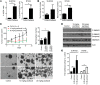
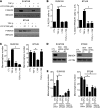
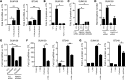
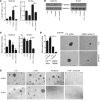
After an initial response to chemotherapy, many patients with triple-negative breast cancer (TNBC) have recurrence of drug-resistant metastatic disease. Studies with TNBC cells suggest that chemotherapy-resistant populations of cancer stem-like cells (CSCs) with self-renewing and tumor-initiating capacities are responsible for these relapses. TGF-β has been shown to increase stem-like properties in human breast cancer cells. We analyzed RNA expression in matched pairs of primary breast cancer biopsies before and after chemotherapy. Biopsies after chemotherapy displayed increased RNA transcripts of genes associated with CSCs and TGF-β signaling. In TNBC cell lines and mouse xenografts, the chemotherapeutic drug paclitaxel increased autocrine TGF-β signaling and IL-8 expression and enriched for CSCs, as indicated by mammosphere formation and CSC markers. The TGF-β type I receptor kinase inhibitor LY2157299, a neutralizing TGF-β type II receptor antibody, and SMAD4 siRNA all blocked paclitaxel-induced IL8 transcription and CSC expansion. Moreover, treatment of TNBC xenografts with LY2157299 prevented reestablishment of tumors after paclitaxel treatment. These data suggest that chemotherapy-induced TGF-β signaling enhances tumor recurrence through IL-8-dependent expansion of CSCs and that TGF-β pathway inhibitors prevent the development of drug-resistant CSCs. These findings support testing a combination of TGF-β inhibitors and anticancer chemotherapy in patients with TNBC.
Figures

Similar articles
-
Transforming growth factor-β signalling controls human breast cancer metastasis in a zebrafish xenograft model.Breast Cancer Res. 2013 Nov 7;15(6):R106. doi: 10.1186/bcr3573. Breast Cancer Res. 2013. PMID: 24196484 Free PMC article.
-
Promoting tumor penetration of nanoparticles for cancer stem cell therapy by TGF-β signaling pathway inhibition.Biomaterials. 2016 Mar;82:48-59. doi: 10.1016/j.biomaterials.2015.12.014. Epub 2015 Dec 21. Biomaterials. 2016. PMID: 26751819
-
An autocrine inflammatory forward-feedback loop after chemotherapy withdrawal facilitates the repopulation of drug-resistant breast cancer cells.Cell Death Dis. 2017 Jul 13;8(7):e2932. doi: 10.1038/cddis.2017.319. Cell Death Dis. 2017. PMID: 28703802 Free PMC article.
-
Clinical development of galunisertib (LY2157299 monohydrate), a small molecule inhibitor of transforming growth factor-beta signaling pathway.Drug Des Devel Ther. 2015 Aug 10;9:4479-99. doi: 10.2147/DDDT.S86621. eCollection 2015. Drug Des Devel Ther. 2015. PMID: 26309397 Free PMC article. Review.
-
Recent advances reveal IL-8 signaling as a potential key to targeting breast cancer stem cells.Breast Cancer Res. 2013;15(4):210. doi: 10.1186/bcr3436. Breast Cancer Res. 2013. PMID: 24041156 Free PMC article. Review.
Cited by
-
SOX4 and SMARCA4 cooperatively regulate PI3k signaling through transcriptional activation of TGFBR2.NPJ Breast Cancer. 2021 Apr 9;7(1):40. doi: 10.1038/s41523-021-00248-2. NPJ Breast Cancer. 2021. PMID: 33837205 Free PMC article.
-
Nanoparticles for Chemoimmunotherapy Against Triple-Negative Breast Cancer.Int J Nanomedicine. 2022 Nov 7;17:5209-5227. doi: 10.2147/IJN.S388075. eCollection 2022. Int J Nanomedicine. 2022. PMID: 36388877 Free PMC article. Review.
-
Molecular alterations and targeted therapy in pancreatic ductal adenocarcinoma.J Hematol Oncol. 2020 Oct 2;13(1):130. doi: 10.1186/s13045-020-00958-3. J Hematol Oncol. 2020. PMID: 33008426 Free PMC article. Review.
-
Detection of phenotype-specific therapeutic vulnerabilities in breast cells using a CRISPR loss-of-function screen.Mol Oncol. 2021 Aug;15(8):2026-2045. doi: 10.1002/1878-0261.12951. Epub 2021 May 1. Mol Oncol. 2021. PMID: 33759347 Free PMC article.
-
A bioinformatic analysis of the inhibin-betaglycan-endoglin/CD105 network reveals prognostic value in multiple solid tumors.PLoS One. 2021 Apr 5;16(4):e0249558. doi: 10.1371/journal.pone.0249558. eCollection 2021. PLoS One. 2021. PMID: 33819300 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

